A novel epimerase that converts GlcNAc-P-P-undecaprenol to GalNAc-P-P-undecaprenol in Escherichia coli O157
- PMID: 19923219
- PMCID: PMC2804325
- DOI: 10.1074/jbc.M109.061630
A novel epimerase that converts GlcNAc-P-P-undecaprenol to GalNAc-P-P-undecaprenol in Escherichia coli O157
Abstract
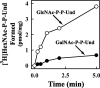
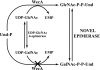
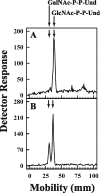
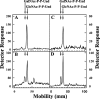
Escherichia coli strain O157 produces an O-antigen with the repeating tetrasaccharide unit alpha-D-PerNAc-alpha-l-Fuc-beta-D-Glc-alpha-D-GalNAc, preassembled on undecaprenyl pyrophosphate (Und-P-P). These studies were conducted to determine whether the biosynthesis of the lipid-linked repeating tetrasaccharide was initiated by the formation of GalNAc-P-P-Und by WecA. When membrane fractions from E. coli strains K12, O157, and PR4019, a WecA-overexpressing strain, were incubated with UDP-[3H]GalNAc, neither the enzymatic synthesis of [3H]GlcNAc-P-P-Und nor [3H]GalNAc-P-P-Und was detected. However, when membrane fractions from strain O157 were incubated with UDP-[3H]GlcNAc, two enzymatically labeled products were observed with the chemical and chromatographic properties of [3H]GlcNAc-P-P-Und and [3H]GalNAc-P-P-Und, suggesting that strain O157 contained an epimerase capable of interconverting GlcNAc-P-P-Und and GalNAc-P-P-Und. The presence of a novel epimerase was demonstrated by showing that exogenous [3H]GlcNAc-P-P-Und was converted to [3H]GalNAc-P-P-Und when incubated with membranes from strain O157. When strain O157 was metabolically labeled with [3H]GlcNAc, both [3H]GlcNAc-P-P-Und and [3H]GalNAc-P-P-Und were detected. Transformation of E. coli strain 21546 with the Z3206 gene enabled these cells to synthesize GalNAc-P-P-Und in vivo and in vitro. The reversibility of the epimerase reaction was demonstrated by showing that [3H]GlcNAc-P-P-Und was reformed when membranes from strain O157 were incubated with exogenous [3H]GalNAc-P-P-Und. The inability of Z3206 to complement the loss of the gne gene in the expression of the Campylobacter jejuni N-glycosylation system in E. coli indicated that it does not function as a UDP-GlcNAc/UDP-GalNAc epimerase. Based on these results, GalNAc-P-P-Und is synthesized reversibly by a novel GlcNAc-P-P-Und epimerase after the formation of GlcNAc-P-P-Und by WecA in E. coli O157.
Figures




References
-
- Whitfield C. (2006) Annu. Rev. Biochem. 75, 39–68 - PubMed
-
- Samuel G., Reeves P. R. (2003) Carbohydr. Res. 338, 2503–2519 - PubMed
-
- Reeves P. R., Hobbs M., Valvano M. A., Skurnik M., Whitfield C., Coplin D., Kido N., Klena J., Maskell D., Raetz C. R., Rick P. D. (1996) Trends Microbiol. 4, 495–503 - PubMed
-
- Law D. (2000) J. Appl. Microbiol. 88, 729–745 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

