GeMMA: functional subfamily classification within superfamilies of predicted protein structural domains
- PMID: 19923231
- PMCID: PMC2817468
- DOI: 10.1093/nar/gkp1049
GeMMA: functional subfamily classification within superfamilies of predicted protein structural domains
Abstract
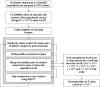
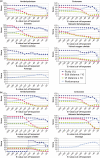
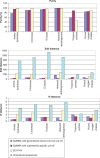
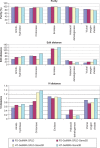
GeMMA (Genome Modelling and Model Annotation) is a new approach to automatic functional subfamily classification within families and superfamilies of protein sequences. A major advantage of GeMMA is its ability to subclassify very large and diverse superfamilies with tens of thousands of members, without the need for an initial multiple sequence alignment. Its performance is shown to be comparable to the established high-performance method SCI-PHY. GeMMA follows an agglomerative clustering protocol that uses existing software for sensitive and accurate multiple sequence alignment and profile-profile comparison. The produced subfamilies are shown to be equivalent in quality whether whole protein sequences are used or just the sequences of component predicted structural domains. A faster, heuristic version of GeMMA that also uses distributed computing is shown to maintain the performance levels of the original implementation. The use of GeMMA to increase the functional annotation coverage of functionally diverse Pfam families is demonstrated. It is further shown how GeMMA clusters can help to predict the impact of experimentally determining a protein domain structure on comparative protein modelling coverage, in the context of structural genomics.
Figures


References
-
- Lee D, Redfern O, Orengo C. Predicting protein function from sequence and structure. Nat. Rev. Mol. Cell Biol. 2007;8:995–1005. - PubMed
-
- Brenner SE. Errors in genome annotation. Trends Genet. 1999;15:132–133. - PubMed
-
- Devos D, Valencia A. Intrinsic errors in genome annotation. Trends Genet. 2001;17:429–431. - PubMed

