A minimal sequence code for switching protein structure and function
- PMID: 19923431
- PMCID: PMC2779201
- DOI: 10.1073/pnas.0906408106
A minimal sequence code for switching protein structure and function
Abstract
We present here a structural and mechanistic description of how a protein changes its fold and function, mutation by mutation. Our approach was to create 2 proteins that (i) are stably folded into 2 different folds, (ii) have 2 different functions, and (iii) are very similar in sequence. In this simplified sequence space we explore the mutational path from one fold to another. We show that an IgG-binding, 4beta+alpha fold can be transformed into an albumin-binding, 3-alpha fold via a mutational pathway in which neither function nor native structure is completely lost. The stabilities of all mutants along the pathway are evaluated, key high-resolution structures are determined by NMR, and an explanation of the switching mechanism is provided. We show that the conformational switch from 4beta+alpha to 3-alpha structure can occur via a single amino acid substitution. On one side of the switch point, the 4beta+alpha fold is >90% populated (pH 7.2, 20 degrees C). A single mutation switches the conformation to the 3-alpha fold, which is >90% populated (pH 7.2, 20 degrees C). We further show that a bifunctional protein exists at the switch point with affinity for both IgG and albumin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


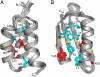
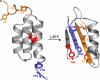
Comment in
-
One sequence plus one mutation equals two folds.Proc Natl Acad Sci U S A. 2009 Dec 15;106(50):21011-2. doi: 10.1073/pnas.0912370107. Epub 2009 Dec 8. Proc Natl Acad Sci U S A. 2009. PMID: 19996167 Free PMC article. No abstract available.
References
-
- Ambroggio XI, Kuhlman B. Design of protein conformational switches. Curr Opin Struct Biol. 2006;16:525–530. - PubMed
-
- Alexander PA, Rozak DA, Orban J, Bryan PN. Directed evolution of highly homologous proteins with different folds by phage display: Implications for the protein folding code. Biochemistry. 2005;44:14045–14054. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

