Rapid universal identification of bacterial pathogens from clinical cultures by using a novel sloppy molecular beacon melting temperature signature technique
- PMID: 19923485
- PMCID: PMC2812257
- DOI: 10.1128/JCM.01725-09
Rapid universal identification of bacterial pathogens from clinical cultures by using a novel sloppy molecular beacon melting temperature signature technique
Abstract
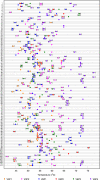
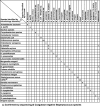
A real-time PCR assay with the ability to rapidly identify all pathogenic bacteria would have widespread medical utility. Current real-time PCR technologies cannot accomplish this task due to severe limitations in multiplexing ability. To this end, we developed a new assay system which supports very high degrees of multiplexing. We developed a new class of mismatch-tolerant "sloppy" molecular beacons, modified them to provide an extended hybridization range, and developed a multiprobe, multimelting temperature (T(m)) signature approach to bacterial species identification. Sloppy molecular beacons were exceptionally versatile, and they were able to generate specific T(m) values for DNA sequences that differed by as little as one nucleotide to as many as 23 polymorphisms. Combining the T(m) values generated by several probe-target hybrids resulted in T(m) signatures that served as highly accurate sequence identifiers. Using this method, PCR assays with as few as six sloppy molecular beacons targeting bacterial 16S rRNA gene segments could reproducibly classify 119 different sequence types of pathogenic and commensal bacteria, representing 64 genera, into 111 T(m) signature types. Blinded studies using the assay to identify the bacteria present in 270 patient-derived clinical cultures including 106 patient blood cultures showed a 95 to 97% concordance with conventional methods. Importantly, no bacteria were misidentified; rather, the few species that could not be identified were classified as "indeterminate," resulting in an assay specificity of 100%. This approach enables highly multiplexed target detection using a simple PCR format that can transform infectious disease diagnostics and improve patient outcomes.
Figures



References
-
- Baldwin, C. D., G. B. Howe, R. Sampath, L. B. Blyn, H. Matthews, V. Harpin, T. A. Hall, J. J. Drader, S. A. Hofstadler, M. W. Eshoo, K. Rudnick, K. Studarus, D. Moore, S. Abbott, J. M. Janda, and C. A. Whitehouse. 2009. Usefulness of multilocus polymerase chain reaction followed by electrospray ionization mass spectrometry to identify a diverse panel of bacterial isolates. Diagn. Microbiol. Infect. Dis. 63:403-408. - PubMed
-
- Bosshard, P. P., R. Zbinden, S. Abels, B. Boddinghaus, M. Altwegg, and E. C. Bottger. 2006. 16S rRNA gene sequencing versus the API 20 NE system and the VITEK 2 ID-GNB card for identification of nonfermenting Gram-negative bacteria in the clinical laboratory. J. Clin. Microbiol. 44:1359-1366. - PMC - PubMed
-
- Claesson, M. J., O. O'Sullivan, Q. Wang, J. Nikkila, J. R. Marchesi, H. Smidt, W. M. de Vos, R. P. Ross, and P. W. O'Toole. 2009. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 4:e6669. - PMC - PubMed
-
- d'Azevedo, P. A., C. A. Dias, A. L. Goncalves, F. Rowe, and L. M. Teixeira. 2001. Evaluation of an automated system for the identification and antimicrobial susceptibility testing of enterococci. Diagn. Microbiol. Infect. Dis. 40:157-161. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

