Structures of BIR domains from human NAIP and cIAP2
- PMID: 19923725
- PMCID: PMC2777033
- DOI: 10.1107/S1744309109038597
Structures of BIR domains from human NAIP and cIAP2
Abstract
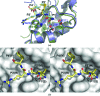
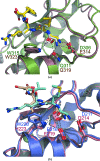
The inhibitor of apoptosis (IAP) family of proteins contains key modulators of apoptosis and inflammation that interact with caspases through baculovirus IAP-repeat (BIR) domains. Overexpression of IAP proteins frequently occurs in cancer cells, thus counteracting the activated apoptotic program. The IAP proteins have therefore emerged as promising targets for cancer therapy. In this work, X-ray crystallography was used to determine the first structures of BIR domains from human NAIP and cIAP2. Both structures harbour an N-terminal tetrapeptide in the conserved peptide-binding groove. The structures reveal that these two proteins bind the tetrapeptides in a similar mode as do other BIR domains. Detailed interactions are described for the P1'-P4' side chains of the peptide, providing a structural basis for peptide-specific recognition. An arginine side chain in the P3' position reveals favourable interactions with its hydrophobic moiety in the binding pocket, while hydrophobic residues in the P2' and P4' pockets make similar interactions to those seen in other BIR domain-peptide complexes. The structures also reveal how a serine in the P1' position is accommodated in the binding pockets of NAIP and cIAP2. In addition to shedding light on the specificity determinants of these two proteins, the structures should now also provide a framework for future structure-based work targeting these proteins.
Figures
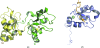

Similar articles
-
Neuronal apoptosis-inhibitory protein does not interact with Smac and requires ATP to bind caspase-9.J Biol Chem. 2004 Sep 24;279(39):40622-8. doi: 10.1074/jbc.M405963200. Epub 2004 Jul 26. J Biol Chem. 2004. PMID: 15280366
-
The mechanism of peptide-binding specificity of IAP BIR domains.Cell Death Differ. 2008 May;15(5):920-8. doi: 10.1038/cdd.2008.6. Epub 2008 Feb 1. Cell Death Differ. 2008. PMID: 18239672
-
Genetic and Epigenetic Regulation of the Innate Immune Response to Gout.Immunol Invest. 2023 Apr;52(3):364-397. doi: 10.1080/08820139.2023.2168554. Epub 2023 Feb 6. Immunol Invest. 2023. PMID: 36745138 Review.
-
Design, synthesis, and biological activity of a potent Smac mimetic that sensitizes cancer cells to apoptosis by antagonizing IAPs.ACS Chem Biol. 2006 Sep 19;1(8):525-33. doi: 10.1021/cb600276q. ACS Chem Biol. 2006. PMID: 17168540
-
IAPs: Modular regulators of cell signalling.Semin Cell Dev Biol. 2015 Mar;39:80-90. doi: 10.1016/j.semcdb.2014.12.002. Epub 2014 Dec 24. Semin Cell Dev Biol. 2015. PMID: 25542341 Review.
Cited by
-
The structure of XIAP BIR2: understanding the selectivity of the BIR domains.Acta Crystallogr D Biol Crystallogr. 2013 Sep;69(Pt 9):1717-25. doi: 10.1107/S0907444913016284. Epub 2013 Aug 15. Acta Crystallogr D Biol Crystallogr. 2013. PMID: 23999295 Free PMC article.
-
Apoptosis inhibition or inflammation: the role of NAIP protein expression in Hodgkin and non-Hodgkin lymphomas compared to non-neoplastic lymph node.J Inflamm (Lond). 2012 Feb 23;9(1):4. doi: 10.1186/1476-9255-9-4. J Inflamm (Lond). 2012. PMID: 22357131 Free PMC article.
-
Inhibitor of apoptosis (IAP)-like protein lacks a baculovirus IAP repeat (BIR) domain and attenuates cell death in plant and animal systems.J Biol Chem. 2011 Dec 9;286(49):42670-42678. doi: 10.1074/jbc.M111.262204. Epub 2011 Sep 16. J Biol Chem. 2011. PMID: 21926169 Free PMC article.
-
Exploration of Key Immune-Related Transcriptomes Associated with Doxorubicin-Induced Cardiotoxicity in Patients with Breast Cancer.Cardiovasc Toxicol. 2023 Oct;23(9-10):329-348. doi: 10.1007/s12012-023-09806-5. Epub 2023 Sep 8. Cardiovasc Toxicol. 2023. PMID: 37684436 Free PMC article.
-
The Combined Use of in Silico, in Vitro, and in Vivo Analyses to Assess Anti-cancerous Potential of a Bioactive Compound from Cyanobacterium Nostoc sp. MGL001.Front Pharmacol. 2017 Nov 27;8:873. doi: 10.3389/fphar.2017.00873. eCollection 2017. Front Pharmacol. 2017. PMID: 29230175 Free PMC article.
References
-
- Chai, J., Shiozaki, E., Srinivasula, S. M., Wu, Q., Datta, P., Alnemri, E. S. & Shi, Y. (2001). Cell, 104, 769–780. - PubMed
-
- Cossu, F., Mastrangelo, E., Milani, M., Sorrentino, G., Lecis, D., Delia, D., Manzoni, L., Seneci, P., Scolastico, C. & Bolognesi, M. (2009). Biochem. Biophys. Res. Commun.378, 162–167. - PubMed
-
- Davoodi, J., Lin, L., Kelly, J., Liston, P. & MacKenzie, A. E. (2004). J. Biol. Chem.279, 40622–40628. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

