New mouse lines for the analysis of neuronal morphology using CreER(T)/loxP-directed sparse labeling
- PMID: 19924248
- PMCID: PMC2775668
- DOI: 10.1371/journal.pone.0007859
New mouse lines for the analysis of neuronal morphology using CreER(T)/loxP-directed sparse labeling
Abstract
Background: Pharmacologic control of Cre-mediated recombination using tamoxifen-dependent activation of a Cre-estrogen receptor ligand binding domain fusion protein [CreER(T)] is widely used to modify and/or visualize cells in the mouse.
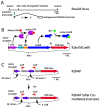
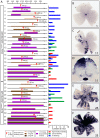
Methods and findings: We describe here two new mouse lines, constructed by gene targeting to the Rosa26 locus to facilitate Cre-mediated cell modification. These lines should prove particularly useful in the context of sparse labeling experiments. The R26rtTACreER line provides ubiquitous expression of CreER under transcriptional control by the tetracycline reverse transactivator (rtTA); dual control by doxycycline and tamoxifen provides an extended dynamic range of Cre-mediated recombination activity. The R26IAP line provides high efficiency Cre-mediated activation of human placental alkaline phosphatase (hPLAP), complementing the widely used, but low efficiency, Z/AP line. By crossing with mouse lines that direct cell-type specific CreER expression, the R26IAP line has been used to produce atlases of labeled cholinergic and catecholaminergic neurons in the mouse brain. The R26IAP line has also been used to visualize the full morphologies of retinal dopaminergic amacrine cells, among the largest neurons in the mammalian retina.
Conclusions: The two new mouse lines described here expand the repertoire of genetically engineered mice available for controlled in vivo recombination and cell labeling using the Cre-lox system.
Conflict of interest statement
Figures





References
-
- Branda CS, Dymecki SM. Talking about a revolution: the impact of site-specific recombinases on genetic analysis in mice. Dev Cell. 2004;6:7–28. - PubMed
-
- Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. - PubMed
-
- Badea T, Nathans J. Quantitative analysis of neuronal morphologies in the mouse retina visualized using a genetically directed reporter. J Comp Neurol. 2004;480:331–351. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

