High resolution cell lineage tracing reveals developmental variability in leech
- PMID: 19924812
- PMCID: PMC3086637
- DOI: 10.1002/dvdy.22158
High resolution cell lineage tracing reveals developmental variability in leech
Abstract
Knowing the normal patterns of embryonic cell proliferation, migration, and differentiation is a cornerstone for understanding development. Yet for most species, the precision with which embryonic cell lineages can be determined is limited by technical considerations (the large numbers of cells, extended developmental times, opacity of the embryos), and these are exacerbated by the inherent variability of the lineages themselves. Here, we present an improved method of cell lineage tracing in the leech Helobdella, driving the expression of a nuclearly localized histone H2B:GFP (green fluorescent protein) fusion protein in selected lineages by microinjection of a plasmid vector. This construct generates a long lasting and minimally mosaic signal with single cell resolution, and does not disrupt the development of most lineages tested. We have validated this technique by elucidating details of cell lineages contributing to segmental and prostomial tissues that could not be observed with standard dextran lineage tracers.
(c) 2009 Wiley-Liss, Inc.
Figures
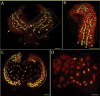

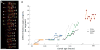
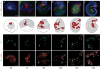

Similar articles
-
Applications of mRNA injections for analyzing cell lineage and asymmetric cell divisions during segmentation in the leech Helobdella robusta.Development. 2005 May;132(9):2103-13. doi: 10.1242/dev.01802. Epub 2005 Mar 23. Development. 2005. PMID: 15788451
-
Grandparental stem cells in leech segmentation: differences in CDC42 expression are correlated with an alternating pattern of blast cell fates.Dev Biol. 2009 Dec 1;336(1):112-21. doi: 10.1016/j.ydbio.2009.09.006. Epub 2009 Sep 9. Dev Biol. 2009. PMID: 19747476 Free PMC article.
-
Asymmetric cell divisions in the early embryo of the leech Helobdella robusta.Prog Mol Subcell Biol. 2007;45:79-95. doi: 10.1007/978-3-540-69161-7_4. Prog Mol Subcell Biol. 2007. PMID: 17585497 Review.
-
An axial domain of HOM/Hox gene expression is formed by morphogenetic alignment of independently specified cell lineages in the leech Helobdella.Development. 1994 Jul;120(7):1839-49. doi: 10.1242/dev.120.7.1839. Development. 1994. PMID: 7924991
-
Developmental control of cell division in leech embryos.Bioessays. 1997 Mar;19(3):201-7. doi: 10.1002/bies.950190305. Bioessays. 1997. PMID: 9080769 Review.
Cited by
-
Pax3/7 regulates neural tube closure and patterning in a non-vertebrate chordate.Front Cell Dev Biol. 2022 Sep 12;10:999511. doi: 10.3389/fcell.2022.999511. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36172287 Free PMC article.
-
Fine taxonomic sampling of nervous systems within Naididae (Annelida: Clitellata) reveals evolutionary lability and revised homologies of annelid neural components.Front Zool. 2015 Apr 18;12:8. doi: 10.1186/s12983-015-0100-6. eCollection 2015. Front Zool. 2015. PMID: 25960761 Free PMC article.
-
Effector gene expression underlying neuron subtype-specific traits in the Motor Ganglion of Ciona.Dev Biol. 2020 Feb 1;458(1):52-63. doi: 10.1016/j.ydbio.2019.10.012. Epub 2019 Oct 19. Dev Biol. 2020. PMID: 31639337 Free PMC article.
-
Cell lineage and cell cycling analyses of the 4d micromere using live imaging in the marine annelid Platynereis dumerilii.Elife. 2017 Dec 12;6:e30463. doi: 10.7554/eLife.30463. Elife. 2017. PMID: 29231816 Free PMC article.
-
Lineage analysis of micromere 4d, a super-phylotypic cell for Lophotrochozoa, in the leech Helobdella and the sludgeworm Tubifex.Dev Biol. 2011 May 1;353(1):120-33. doi: 10.1016/j.ydbio.2011.01.031. Epub 2011 Feb 3. Dev Biol. 2011. PMID: 21295566 Free PMC article.
References
-
- Baker MW, Macagno ER. Characterizations of Hirudo medicinalis DNA promoters for targeted gene expression. J Neurosci Methods. 2006;156:145–153. - PubMed
-
- Baker MW, Macagno ER. In vivo imaging of growth cone and filopodial dynamics: evidence for contact-mediated retraction of filopodia leading to the tiling of sibling processes. J Comp Neurol. 2007;500:850–862. - PubMed
-
- Baker MW, Peterson SM, Macagno ER. The receptor phosphatase HmLAR2 collaborates with focal adhesion proteins in filopodial tips to control growth cone morphology. Dev Biol. 2008;320:215–225. - PubMed
-
- Bissen ST, Weisblat DA. The durations and compositions of cell cycles in embryos of the leech, Helobdella triserialis. Development. 1989;106:105–118. - PubMed
-
- Boyer BC, Henry JQ, Martindale MQ. Dual origins of mesoderm in a basal spiralian: cell lineage analyses in the polyclad turbellarian Hoploplana inquilina. Developmental Biology. 1996;179:329–338. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

