Diversification of the cullin family
- PMID: 19925652
- PMCID: PMC2785787
- DOI: 10.1186/1471-2148-9-267
Diversification of the cullin family
Abstract
Background: Cullins are proteins involved in ubiquitination through their participation in multisubunit ubiquitin ligase complexes. In this study, I use comparative genomic data to establish the pattern of emergence and diversification of cullins in eukaryotes.
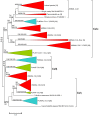
Results: The available data indicate that there were three cullin genes before the unikont/bikont split, which I have called Culalpha, Culbeta and Culgamma. Fungal species have quite strictly conserved these three ancestral genes, with only occasional lineage-specific duplications. On the contrary, several additional genes appeared in the animal or plant lineages. For example, the human genes Cul1, Cul2, Cul5, Cul7 and Parc all derive from the ancestral Culalpha gene. These results, together with the available functional data, suggest that three different types of ubiquitin ligase cullin-containing complexes were already present in early eukaryotic evolution: 1) SCF-like complexes with Culalpha proteins; 2) Culbeta/BTB complexes; and, 3) Complexes containing Culgamma and DDB1-like proteins. Complexes containing elongins have arisen more recently and perhaps twice independently in animals and fungi.
Conclusion: Most of the known types of cullin-containing ubiquitin ligase complexes are ancient. The available data suggest that, since the origin of eukaryotes, complex diversity has been mostly generated by combining closely related subunits, while radical innovations, giving rise to novel types of complexes, have been scarce. However, several protist groups not examined so far contain highly divergent cullins, indicating that additional types of complexes may exist.
Figures




References
-
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. - PubMed

