Organization, structure, and assembly of alpha-carboxysomes determined by electron cryotomography of intact cells
- PMID: 19925807
- PMCID: PMC2853366
- DOI: 10.1016/j.jmb.2009.11.019
Organization, structure, and assembly of alpha-carboxysomes determined by electron cryotomography of intact cells
Abstract
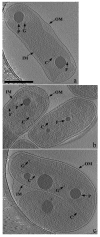
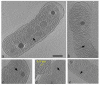
Carboxysomes are polyhedral inclusion bodies that play a key role in autotrophic metabolism in many bacteria. Using electron cryotomography, we examined carboxysomes in their native states within intact cells of three chemolithoautotrophic bacteria. We found that carboxysomes generally cluster into distinct groups within the cytoplasm, often in the immediate vicinity of polyphosphate granules, and a regular lattice of density frequently connects granules to nearby carboxysomes. Small granular bodies were also seen within carboxysomes. These observations suggest a functional relationship between carboxysomes and polyphosphate granules. Carboxysomes exhibited greater size, shape, and compositional variability in cells than in purified preparations. Finally, we observed carboxysomes in various stages of assembly, as well as filamentous structures that we attribute to misassembled shell protein. Surprisingly, no more than one partial carboxysome was ever observed per cell. Based on these observations, we propose a model for carboxysome assembly in which the shell and the internal RuBisCO (ribulose-1,5-bisphosphate carboxylase/oxygenase) lattice form simultaneously, likely guided by specific interactions between shell proteins and RuBisCOs.
Copyright 2009 Elsevier Ltd. All rights reserved.
Figures








References
-
- Yeates T, Kerfeld C, Heinhorst S, Cannon GC, Shively JM. Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nat Rev Microbiol. 2008;6:681–691. - PubMed
-
- Badger MR, Price GD. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. Journal of Experimental Botany. 2003;54:609–622. - PubMed
-
- Spreitzer RJ, Salvucci ME. Rubisco: structure, regulatory interactions, and possibilities for a better enzyme. Annual review of plant biology. 2002;53:449–75. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

