Genetic resources for maize cell wall biology
- PMID: 19926802
- PMCID: PMC2785990
- DOI: 10.1104/pp.109.136804
Genetic resources for maize cell wall biology
Abstract
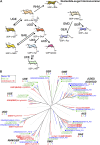
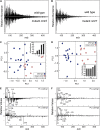
Grass species represent a major source of food, feed, and fiber crops and potential feedstocks for biofuel production. Most of the biomass is contributed by cell walls that are distinct in composition from all other flowering plants. Identifying cell wall-related genes and their functions underpins a fundamental understanding of growth and development in these species. Toward this goal, we are building a knowledge base of the maize (Zea mays) genes involved in cell wall biology, their expression profiles, and the phenotypic consequences of mutation. Over 750 maize genes were annotated and assembled into gene families predicted to function in cell wall biogenesis. Comparative genomics of maize, rice (Oryza sativa), and Arabidopsis (Arabidopsis thaliana) sequences reveal differences in gene family structure between grass species and a reference eudicot species. Analysis of transcript profile data for cell wall genes in developing maize ovaries revealed that expression within families differed by up to 100-fold. When transcriptional analyses of developing ovaries before pollination from Arabidopsis, rice, and maize were contrasted, distinct sets of cell wall genes were expressed in grasses. These differences in gene family structure and expression between Arabidopsis and the grasses underscore the requirement for a grass-specific genetic model for functional analyses. A UniformMu population proved to be an important resource in both forward- and reverse-genetics approaches to identify hundreds of mutants in cell wall genes. A forward screen of field-grown lines by near-infrared spectroscopic screen of mature leaves yielded several dozen lines with heritable spectroscopic phenotypes. Pyrolysis-molecular beam mass spectrometry confirmed that several nir mutants had altered carbohydrate-lignin compositions.
Figures










References
-
- Ahn JW, Verma R, Kim M, Lee JY, Kim YK, Bang JW, Reiter WD, Pal HS (2006) Depletion of UDP-D-apiose/UDP-D-xylose synthases results in rhamnogalacturonan II deficiency, cell wall thickening, and cell death in higher plants. J Biol Chem 281 13708–13716 - PubMed
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215 403–410 - PubMed
-
- Appenzeller L, Doblin M, Barreiro R, Wang HY, Niu XM, Kollipara K, Carrigan L, Tomes D, Chapman M, Dhugga KS (2004) Cellulose synthesis in maize: isolation and expression analysis of the cellulose synthase (CesA) gene family. Cellulose 11 287–299
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

