Design of O-acetylserine sulfhydrylase inhibitors by mimicking nature
- PMID: 19928859
- PMCID: PMC2804909
- DOI: 10.1021/jm901325e
Design of O-acetylserine sulfhydrylase inhibitors by mimicking nature
Abstract
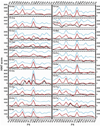
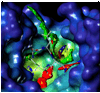
The inhibition of cysteine biosynthesis in prokaryotes and protozoa has been proposed to be relevant for the development of antibiotics. Haemophilus influenzae O-acetylserine sulfhydrylase (OASS), catalyzing l-cysteine formation, is inhibited by the insertion of the C-terminal pentapeptide (MNLNI) of serine acetyltransferase into the active site. Four-hundred MNXXI pentapeptides were generated in silico, docked into OASS active site using GOLD, and scored with HINT. The terminal P5 Ile accounts for about 50% of the binding energy. Glu or Asp at position P4 and, to a lesser extent, at position P3 also significantly contribute to the binding interaction. The predicted affinity of 14 selected pentapeptides correlated well with the experimentally determined dissociation constants. The X-ray structure of three high affinity pentapeptide-OASS complexes were compared with the docked poses. These results, combined with a GRID analysis of the active site, allowed us to define a pharmacophoric scaffold for the design of peptidomimetic inhibitors.
Figures






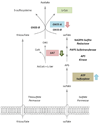
 ) and activation (
) and activation ( ) of enzyme activities are shown in colour. Cysteine inhibits SAT and SAT inhibits OASS-A. OASS-A activates ATP sulfurylase.
) of enzyme activities are shown in colour. Cysteine inhibits SAT and SAT inhibits OASS-A. OASS-A activates ATP sulfurylase.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Chemical Information
Miscellaneous

