A whole genome Bayesian scan for adaptive genetic divergence in West African cattle
- PMID: 19930592
- PMCID: PMC2784811
- DOI: 10.1186/1471-2164-10-550
A whole genome Bayesian scan for adaptive genetic divergence in West African cattle
Abstract
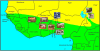
Background: The recent settlement of cattle in West Africa after several waves of migration from remote centres of domestication has imposed dramatic changes in their environmental conditions, in particular through exposure to new pathogens. West African cattle populations thus represent an appealing model to unravel the genome response to adaptation to tropical conditions. The purpose of this study was to identify footprints of adaptive selection at the whole genome level in a newly collected data set comprising 36,320 SNPs genotyped in 9 West African cattle populations.
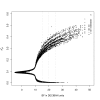
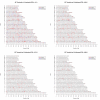
Results: After a detailed analysis of population structure, we performed a scan for SNP differentiation via a previously proposed Bayesian procedure including extensions to improve the detection of loci under selection. Based on these results we identified 53 genomic regions and 42 strong candidate genes. Their physiological functions were mainly related to immune response (MHC region which was found under strong balancing selection, CD79A, CXCR4, DLK1, RFX3, SEMA4A, TICAM1 and TRIM21), nervous system (NEUROD6, OLFM2, MAGI1, SEMA4A and HTR4) and skin and hair properties (EDNRB, TRSP1 and KRTAP8-1).
Conclusion: The main possible underlying selective pressures may be related to climatic conditions but also to the host response to pathogens such as Trypanosoma(sp). Overall, these results might open the way towards the identification of important variants involved in adaptation to tropical conditions and in particular to resistance to tropical infectious diseases.
Figures



References
-
- Epstein H. The Origins of the Domestic Animals of Africa. Vol. 1. London: Africana Publishing Corporation; 1971.
-
- Payne WJA, Hodges J. Tropical Cattle, Origins, Breeds and Breeding Policies. Oxford: Blackwell Science Ltd; 1997.
-
- Williamson G, Payne WJA. An introduction to Animal Husbandry in the Tropics. English Language Book Service/Longman; 1984.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

