From where did the 2009 'swine-origin' influenza A virus (H1N1) emerge?
- PMID: 19930669
- PMCID: PMC2787513
- DOI: 10.1186/1743-422X-6-207
From where did the 2009 'swine-origin' influenza A virus (H1N1) emerge?
Abstract
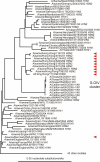
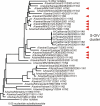
The swine-origin influenza A (H1N1) virus that appeared in 2009 and was first found in human beings in Mexico, is a reassortant with at least three parents. Six of the genes are closest in sequence to those of H1N2 'triple-reassortant' influenza viruses isolated from pigs in North America around 1999-2000. Its other two genes are from different Eurasian 'avian-like' viruses of pigs; the NA gene is closest to H1N1 viruses isolated in Europe in 1991-1993, and the MP gene is closest to H3N2 viruses isolated in Asia in 1999-2000. The sequences of these genes do not directly reveal the immediate source of the virus as the closest were from isolates collected more than a decade before the human pandemic started. The three parents of the virus may have been assembled in one place by natural means, such as by migrating birds, however the consistent link with pig viruses suggests that human activity was involved. We discuss a published suggestion that unsampled pig herds, the intercontinental live pig trade, together with porous quarantine barriers, generated the reassortant. We contrast that suggestion with the possibility that laboratory errors involving the sharing of virus isolates and cultured cells, or perhaps vaccine production, may have been involved. Gene sequences from isolates that bridge the time and phylogenetic gap between the new virus and its parents will distinguish between these possibilities, and we suggest where they should be sought. It is important that the source of the new virus be found if we wish to avoid future pandemics rather than just trying to minimize the consequences after they have emerged. Influenza virus is a very significant zoonotic pathogen. Public confidence in influenza research, and the agribusinesses that are based on influenza's many hosts, has been eroded by several recent events involving the virus. Measures that might restore confidence include establishing a unified international administrative framework coordinating surveillance, research and commercial work with this virus, and maintaining a registry of all influenza isolates.
Figures


References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

