Simultaneous genotype calling and haplotype phasing improves genotype accuracy and reduces false-positive associations for genome-wide association studies
- PMID: 19931040
- PMCID: PMC2790566
- DOI: 10.1016/j.ajhg.2009.11.004
Simultaneous genotype calling and haplotype phasing improves genotype accuracy and reduces false-positive associations for genome-wide association studies
Abstract
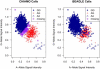
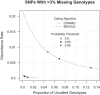
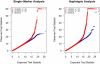
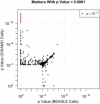
We present a novel method for simultaneous genotype calling and haplotype-phase inference. Our method employs the computationally efficient BEAGLE haplotype-frequency model, which can be applied to large-scale studies with millions of markers and thousands of samples. We compare genotype calls made with our method to genotype calls made with the BIRDSEED, CHIAMO, GenCall, and ILLUMINUS genotype-calling methods, using genotype data from the Illumina 550K and Affymetrix 500K arrays. We show that our method has higher genotype-call accuracy and yields fewer uncalled genotypes than competing methods. We perform single-marker analysis of data from the Wellcome Trust Case Control Consortium bipolar disorder and type 2 diabetes studies. For bipolar disorder, the genotype calls in the original study yield 25 markers with apparent false-positive association with bipolar disorder at a p < 10(-7) significance level, whereas genotype calls made with our method yield no associated markers at this significance threshold. Conversely, for markers with replicated association with type 2 diabetes, there is good concordance between genotype calls used in the original study and calls made by our method. Results from single-marker and haplotypic analysis of our method's genotype calls for the bipolar disorder study indicate that our method is highly effective at eliminating genotyping artifacts that cause false-positive associations in genome-wide association studies. Our new genotype-calling methods are implemented in the BEAGLE and BEAGLECALL software packages.
Figures


References
-
- Frayling T.M. Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat. Rev. Genet. 2007;8:657–662. - PubMed
-
- Zeggini E., Scott L.J., Saxena R., Voight B.F., Marchini J.L., Hu T., de Bakker P.I., Abecasis G.R., Almgren P., Andersen G. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 2008;40:638–645. - PMC - PubMed
-
- Rioux J.D., Xavier R.J., Taylor K.D., Silverberg M.S., Goyette P., Huett A., Green T., Kuballa P., Barmada M.M., Datta L.W. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 2007;39:596–604. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

