An RNA structural switch regulates diploid genome packaging by Moloney murine leukemia virus
- PMID: 19931283
- PMCID: PMC2836482
- DOI: 10.1016/j.jmb.2009.11.033
An RNA structural switch regulates diploid genome packaging by Moloney murine leukemia virus
Abstract
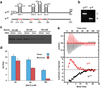
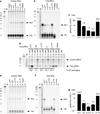
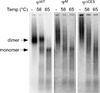
Retroviruses selectively package two copies of their RNA genomes via mechanisms that have yet to be fully deciphered. Recent studies with small fragments of the Moloney murine leukemia virus (MoMuLV) genome suggested that selection may be mediated by an RNA switch mechanism, in which conserved UCUG elements that are sequestered by base-pairing in the monomeric RNA become exposed upon dimerization to allow binding to the cognate nucleocapsid (NC) domains of the viral Gag proteins. Here we show that a large fragment of the MoMuLV 5' untranslated region that contains all residues necessary for efficient RNA packaging (Psi(WT); residues 147-623) also exhibits a dimerization-dependent affinity for NC, with the native dimer ([Psi(WT)](2)) binding 12+/-2 NC molecules with high affinity (K(d)=17+/-7 nM) and with the monomer, stabilized by substitution of dimer-promoting loop residues with hairpin-stabilizing sequences (Psi(M)), binding 1-2 NC molecules. Identical dimer-inhibiting mutations in MoMuLV-based vectors significantly inhibit genome packaging in vivo (approximately 100-fold decrease), whereas a large deletion of nearly 200 nucleotides just upstream of the gag start codon has minimal effects. Our findings support the proposed RNA switch mechanism and further suggest that virus assembly may be initiated by a complex comprising as few as 12 Gag molecules bound to a dimeric packaging signal.
Copyright 2009 Elsevier Ltd. All rights reserved.
Figures

Similar articles
-
Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus.Nature. 2004 Sep 30;431(7008):586-90. doi: 10.1038/nature02944. Nature. 2004. PMID: 15457265
-
Identification of a high affinity nucleocapsid protein binding element within the Moloney murine leukemia virus Psi-RNA packaging signal: implications for genome recognition.J Mol Biol. 2001 Nov 23;314(2):217-32. doi: 10.1006/jmbi.2001.5139. J Mol Biol. 2001. PMID: 11718556
-
Composition and sequence-dependent binding of RNA to the nucleocapsid protein of Moloney murine leukemia virus.Biochemistry. 2005 Mar 15;44(10):3735-44. doi: 10.1021/bi047639q. Biochemistry. 2005. PMID: 15751950
-
How retroviruses select their genomes.Nat Rev Microbiol. 2005 Aug;3(8):643-55. doi: 10.1038/nrmicro1210. Nat Rev Microbiol. 2005. PMID: 16064056 Review.
-
Structural determinants and mechanism of HIV-1 genome packaging.J Mol Biol. 2011 Jul 22;410(4):609-33. doi: 10.1016/j.jmb.2011.04.029. J Mol Biol. 2011. PMID: 21762803 Free PMC article. Review.
Cited by
-
A purine loop and the primer binding site are critical for the selective encapsidation of mouse mammary tumor virus genomic RNA by Pr77Gag.Nucleic Acids Res. 2021 May 7;49(8):4668-4688. doi: 10.1093/nar/gkab223. Nucleic Acids Res. 2021. PMID: 33836091 Free PMC article.
-
Selection of RNAs for constructing "Lighting-UP" biomolecular switches in response to specific small molecules.PLoS One. 2013;8(3):e60222. doi: 10.1371/journal.pone.0060222. Epub 2013 Mar 26. PLoS One. 2013. PMID: 23555931 Free PMC article.
-
Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography.J Mol Biol. 2010 Dec 17;404(5):751-72. doi: 10.1016/j.jmb.2010.09.009. Epub 2010 Oct 8. J Mol Biol. 2010. PMID: 20933521 Free PMC article.
-
Structural dynamics of retroviral genome and the packaging.Front Microbiol. 2011 Dec 29;2:264. doi: 10.3389/fmicb.2011.00264. eCollection 2011. Front Microbiol. 2011. PMID: 22232618 Free PMC article.
-
Insights into the nuclear export of murine leukemia virus intron-containing RNA.RNA Biol. 2015;12(9):942-9. doi: 10.1080/15476286.2015.1065375. RNA Biol. 2015. PMID: 26158194 Free PMC article. Review.
References
-
- Berkowitz R, Fisher J, Goff SP. RNA packaging. Curr. Top. Microbiol. Immun. 1996;214:177–218. - PubMed
-
- Varmus HE. Form and function of retroviral proviruses. Science. 1982;216:812–820. - PubMed
-
- Hu WS, Temin HM. Retroviral recombination and reverse transcription. Science. 1990;250:1227–1233. - PubMed
-
- Rein A. Retroviral RNA packaging: A review. Arch. Virology. 1994;9:513–522. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

