Visualization of individual Scr mRNAs during Drosophila embryogenesis yields evidence for transcriptional bursting
- PMID: 19931455
- PMCID: PMC2805773
- DOI: 10.1016/j.cub.2009.10.028
Visualization of individual Scr mRNAs during Drosophila embryogenesis yields evidence for transcriptional bursting
Abstract
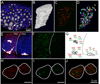
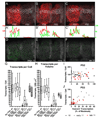
The detection and counting of transcripts within single cells via fluorescent in situ hybridization (FISH) has allowed researchers to ask quantitative questions about gene expression at the level of individual cells. This method is often preferable to quantitative RT-PCR, because it does not necessitate destruction of the cells being probed and maintains spatial information that may be of interest. Until now, studies using FISH at single-molecule resolution have only been rigorously carried out in isolated cells (e.g., yeast cells or mammalian cell culture). Here, we describe the detection and counting of transcripts within single cells of fixed, whole-mount Drosophila embryos via a combination of FISH, immunohistochemistry, and image segmentation. Our method takes advantage of inexpensive, long RNA probes detected with antibodies, and we present novel evidence to show that we can robustly detect single mRNA molecules. We use this method to characterize transcription at the endogenous locus of the Hox gene Sex combs reduced (Scr), by comparing a stably expressing group of cells to a group that only transiently expresses the gene. Our data provide evidence for transcriptional bursting, as well for divergent "accumulation" and "maintenance" phases of gene activity at the Scr locus.
Figures


References
-
- Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science (New York, N.Y.) 1998;280:585–590. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

