Do all modifications benefit all tRNAs?
- PMID: 19931536
- PMCID: PMC2794898
- DOI: 10.1016/j.febslet.2009.11.049
Do all modifications benefit all tRNAs?
Abstract
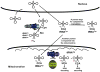
Despite the universality of tRNA modifications, some tRNAs lacking specific modifications are subject to degradation pathways, while other tRNAs lacking the same modifications are resistant. Here, we suggest a model in which some modifications have minor, possibly redundant, roles in specific tRNAs. This model is consistent with the low specificity of some modification enzymes. Limitations of this model include the limited assays and growth conditions on which these conclusions are based, as well as the high specificity exhibited by many modification enzymes with important roles in translation. The specificity of these enzymes is often enhanced by complex substrate recognition patterns and sub-cellular compartmentalization.
Figures
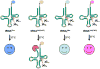
References
-
- Fraser CM, et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. - PubMed
-
- Noon KR, Guymon R, Crain PF, McCloskey JA, Thomm M, Lim J, Cavicchioli R. Influence of temperature on tRNA modification in archaea: Methanococcoides burtonii (optimum growth temperature [Topt], 23 degrees C) and Stetteria hydrogenophila (Topt, 95 degrees C) J Bacteriol. 2003;185:5483–90. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

