Nonallelic transvection of multiple imprinted loci is organized by the H19 imprinting control region during germline development
- PMID: 19933149
- PMCID: PMC2779760
- DOI: 10.1101/gad.552109
Nonallelic transvection of multiple imprinted loci is organized by the H19 imprinting control region during germline development
Abstract
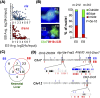
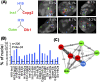
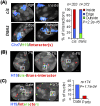
Recent observations highlight that the mammalian genome extensively communicates with itself via long-range chromatin interactions. The causal link between such chromatin cross-talk and epigenetic states is, however, poorly understood. We identify here a network of physically juxtaposed regions from the entire genome with the common denominator of being genomically imprinted. Moreover, CTCF-binding sites within the H19 imprinting control region (ICR) not only determine the physical proximity among imprinted domains, but also transvect allele-specific epigenetic states, identified by replication timing patterns, to interacting, nonallelic imprinted regions during germline development. We conclude that one locus can directly or indirectly pleiotropically influence epigenetic states of multiple regions on other chromosomes with which it interacts.
Figures

References
-
- Bergstrom R, Whitehead J, Kurukuti S, Ohlsson R. CTCF regulates asynchronous replication of the imprinted H19/Igf2 domain. Cell Cycle. 2007;6:450–454. - PubMed
-
- Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet. 2006;38:1278–1288. - PubMed
-
- Cerrato F, Dean W, Davies K, Kagotani K, Mitsuya K, Okumura K, Riccio A, Reik W. Paternal imprints can be established on the maternal Igf2-H19 locus without altering replication timing of DNA. Hum Mol Genet. 2003;12:3123–3132. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
