Verification of specific G-quadruplex structure by using a novel cyanine dye supramolecular assembly: II. The binding characterization with specific intramolecular G-quadruplex and the recognizing mechanism
- PMID: 19933263
- PMCID: PMC2817466
- DOI: 10.1093/nar/gkp1045
Verification of specific G-quadruplex structure by using a novel cyanine dye supramolecular assembly: II. The binding characterization with specific intramolecular G-quadruplex and the recognizing mechanism
Abstract
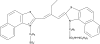
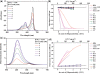
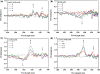
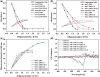
The supramolecular assembly of a novel cyanine dye, 3,3'-di(3-sulfopropyl)-4,5,4',5'-dibenzo-9-ethyl-thiacarbocyanine triethylammonium salt (ETC) was designed to verify specific intramolecular G-quadruplexes from duplex and single-strand DNAs. Spectral results have shown that ETC presented two major distinct signatures with specific intramolecular G-quadruplexes in vitro: (i) dramatic changes in the absorption spectra (including disappearance of absorption peak around 660 nm and appearance of independent new peak around 584 nm); (ii) approximately 70 times enhancement of fluorescence signal at 600 nm. Furthermore, based on (1)H-nuclear magnetic resonance and circular dichroism results, the preferring binding of ETC to specific intramolecular G-quadruplexes probably result from end-stacking, and the loop structure nearby also plays an important role.
Figures

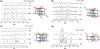
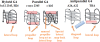


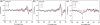

References
-
- Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–573. - PubMed
-
- Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. - PubMed
-
- Pennisi E. Genetics – DNA's; molecular gymnastics. Science. 2006;312:1467–1468. - PubMed
-
- Fu BQ, Huang J, Ren L, Weng XC, Zhou YY, Du YH, Wu XJ, Zhou X, Yang GF. Cationic corrole derivatives: a new family of G-quadruplex inducing and stabilizing ligands. Chem. Commun. 2007:3264–3266. - PubMed
-
- Zhou Q, Li L, Xiang J, Tang Y, Zhang H, Yang S, Li Q, Yang Q, Xu G. Screening potential antitumor agents from natural plant extracts by G-quadruplex recognition and NMR methods. Angew. Chem.-Int. Edit. 2008;47:5590–5592. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

