Improved free energy parameters for RNA pseudoknotted secondary structure prediction
- PMID: 19933322
- PMCID: PMC2802035
- DOI: 10.1261/rna.1689910
Improved free energy parameters for RNA pseudoknotted secondary structure prediction
Abstract
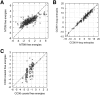
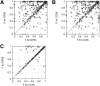
Accurate prediction of RNA pseudoknotted secondary structures from the base sequence is a challenging computational problem. Since prediction algorithms rely on thermodynamic energy models to identify low-energy structures, prediction accuracy relies in large part on the quality of free energy change parameters. In this work, we use our earlier constraint generation and Boltzmann likelihood parameter estimation methods to obtain new energy parameters for two energy models for secondary structures with pseudoknots, namely, the Dirks-Pierce (DP) and the Cao-Chen (CC) models. To train our parameters, and also to test their accuracy, we create a large data set of both pseudoknotted and pseudoknot-free secondary structures. In addition to structural data our training data set also includes thermodynamic data, for which experimentally determined free energy changes are available for sequences and their reference structures. When incorporated into the HotKnots prediction algorithm, our new parameters result in significantly improved secondary structure prediction on our test data set. Specifically, the prediction accuracy when using our new parameters improves from 68% to 79% for the DP model, and from 70% to 77% for the CC model.
Figures
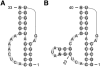
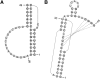
References
-
- Andronescu M. University of British Columbia; Vancouver, Canada: 2003. “Algorithms for predicting the secondary structure of pairs and combinatorial sets of nucleic acid strands.”. MS thesis,
-
- Andronescu MS. University of British Columbia; Vancouver, Canada: 2008. “Computational approaches for RNA energy parameter estimation.”. PhD thesis,
-
- Andronescu M, Condon A, Hoos HH, Mathews DH, Murphy KP. Efficient parameter estimation for RNA secondary structure prediction. Bioinformatics. 2007;23:19–28. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
