Estrogen and xenoestrogens in breast cancer
- PMID: 19933552
- PMCID: PMC2907875
- DOI: 10.1177/0192623309354108
Estrogen and xenoestrogens in breast cancer
Abstract
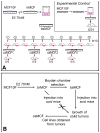
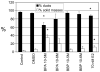
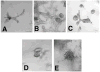
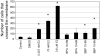
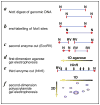
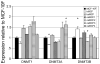
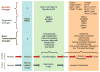
There is growing concern that estrogenic environmental compounds that act as endocrine-disrupting chemicals might potentially have adverse effects on hormone-sensitive organs such as the breast. This concern is further fueled by evidence indicating that natural estrogens, specifically 17beta-estradiol, are important factors in the initiation and progression of breast cancer. We have developed an in vitro-in vivo model in which we have demonstrated the carcinogenicity of E2 in human breast epithelial cells MCF-10F. Hypermethylation of NRG1, STXBP6, BMP6, CSS3, SPRY1, and SNIP were found at different progression stages in this model. The use of this powerful and unique model has provided a tool for exploring whether bisphenol A and butyl benzyl phthalate have relevance in the initiation of breast cancer. These studies provide firsthand evidence that the natural estrogen 17beta-estradiol and xenoestrogenic substances like bisphenol A are able to induce neoplastic transformation in human breast epithelial cells.
Figures


References
-
- Breast cancer and hormone replacement therapy: collaborative reanalysis of data from 51 epidemiological studies of 52,705 women with breast cancer and 108,411 women without breast cancer. Collaborative Group on Hormonal Factors in Breast Cancer. Lancet. 350(9084):1047–59. - PubMed
-
- Akingbemi BT, Sottas CM, Koulova AI, Klinefelter GR, Hardy MP. Inhibition of testicular steroidogenesis by the xenoestrogen bisphenol A is associated with reduced pituitary luteinizing hormone secretion and decreased steroidogenic enzyme gene expression in rat Leydig cells. Endocrinology. 2004;145(2):592–603. - PubMed
-
- Bakken K, Alsaker E, Eggen AE, Lund E. Hormone replacement therapy and incidence of hormone-dependent cancers in the Norwegian Women and Cancer study. Int J Cancer. 2004;112(1):130–4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

