Cdk2 and Cdk4 regulate the centrosome cycle and are critical mediators of centrosome amplification in p53-null cells
- PMID: 19933848
- PMCID: PMC2812235
- DOI: 10.1128/MCB.00253-09
Cdk2 and Cdk4 regulate the centrosome cycle and are critical mediators of centrosome amplification in p53-null cells
Erratum in
-
Correction for Adon et al., "Cdk2 and Cdk4 Regulate the Centrosome Cycle and Are Critical Mediators of Centrosome Amplification in p53-Null Cells".Mol Cell Biol. 2020 Nov 6;40(23):e00488-20. doi: 10.1128/MCB.00488-20. Print 2020 Nov 6. Mol Cell Biol. 2020. PMID: 33159003 Free PMC article. No abstract available.
Abstract
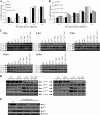
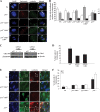
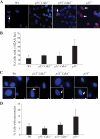
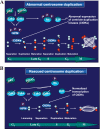
The two mitotic centrosomes direct spindle bipolarity to maintain euploidy. Centrosome amplification-the acquisition of > or =3 centrosomes-generates multipolar mitoses, aneuploidy, and chromosome instability to promote cancer biogenesis. While much evidence suggests that Cdk2 is the major conductor of the centrosome cycle and that it mediates centrosome amplification induced by various altered tumor suppressors, the role played by Cdk4 in a normal or deregulated centrosome cycle is unknown. Using a gene knockout approach, we report that Cdk2 and Cdk4 are critical to the centrosome cycle, since centrosome separation and duplication are premature in Cdk2(-)(/)(-) mouse embryonic fibroblasts (MEFs) and are compromised in Cdk4(-)(/)(-) MEFs. Additionally, ablation of Cdk4 or Cdk2 abrogates centrosome amplification and chromosome instability in p53-null MEFs. Absence of Cdk2 or Cdk4 prevents centrosome amplification by abrogating excessive centriole duplication. Furthermore, hyperactive Cdk2 and Cdk4 deregulate the licensing of the centrosome duplication cycle in p53-null cells by hyperphosphorylating nucleophosmin (NPM) at Thr199, as evidenced by observations that ablation of Cdk2, Cdk4, or both Cdk2 and Cdk4 abrogates that excessive phosphorylation. Since a mutant form of NPM lacking the G(1) Cdk phosphorylation site (NPM(T199A)) prevents centrosome amplification to the same extent as ablation of Cdk2 or Cdk4, we conclude that the Cdk2/Cdk4/NPM pathway is a major guardian of centrosome dysfunction and genomic integrity.
Figures




References
-
- Albertson, D. G., C. Collins, F. McCormick, and J. W. Gray. 2003. Chromosome aberrations in solid tumors. Nat. Genet. 34:369-376. - PubMed
-
- Aleem, E., H. Kiyokawa, and P. Kaldis. 2005. Cdc2-cyclin E complexes regulate the G1/S phase transition. Nat. Cell Biol. 7:831-836. - PubMed
-
- Ball, K. L., S. Lain, R. Fahraeus, C. Smythe, and D. P. Lane. 1997. Cell-cycle arrest and inhibition of Cdk4 activity by small peptides based on the carboxy-terminal domain of p21WAF1. Curr. Biol. 7:71-80. - PubMed
-
- Bearss, D. J., R. J. Lee, D. A. Troyer, R. G. Pestell, and J. J. Windle. 2002. Differential effects of p21(WAF1/CIP1) deficiency on MMTV-ras and MMTV-myc mammary tumor properties. Cancer Res. 62:2077-2084. - PubMed
-
- Berthet, C., E. Aleem, V. Coppola, L. Tessarollo, and P. Kaldis. 2003. Cdk2 knockout mice are viable. Curr. Biol. 13:1775-1785. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
