Coalescent simulation of intracodon recombination
- PMID: 19933876
- PMCID: PMC2828723
- DOI: 10.1534/genetics.109.109736
Coalescent simulation of intracodon recombination
Abstract
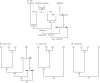
The coalescent with recombination is a very useful tool in molecular population genetics. Under this framework, genealogies often represent the evolution of the substitution unit, and because of this, the few coalescent algorithms implemented for the simulation of coding sequences force recombination to occur only between codons. However, it is clear that recombination is expected to occur most often within codons. Here we have developed an algorithm that can evolve coding sequences under an ancestral recombination graph that represents the genealogies at each nucleotide site, thereby allowing for intracodon recombination. The algorithm is a modification of Hudson's coalescent in which, in addition to keeping track of events occurring in the ancestral material that reaches the sample, we need to keep track of events occurring in ancestral material that does not reach the sample but that is produced by intracodon recombination. We are able to show that at typical substitution rates the number of nonsynonymous changes induced by intracodon recombination is small and that intracodon recombination does not generally result in inflated estimates of the overall nonsynonymous/synonymous substitution ratio (omega). On the other hand, recombination can bias the estimation of omega at particular codons, resulting in apparent rate variation among sites and in the spurious identification of positively selected sites. Importantly, in this case, allowing for variable synonymous rates across sites greatly reduces the false-positive rate and recovers statistical power. Finally, coalescent simulations with intracodon recombination could be used to better represent the evolution of nuclear coding genes or fast-evolving pathogens such as HIV-1.We have implemented this algorithm in a computer program called NetRecodon, freely available at http://darwin.uvigo.es.
Figures



References
-
- Bustamante, C. D., A. Fledel-Alon, S. Williamson, R. Nielsen, M. T. Hubisz et al., 2005. Natural selection on protein-coding genes in the human genome. Nature 437 1153–1157. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

