Cytokines interleukin-1beta and tumor necrosis factor-alpha regulate different transcriptional and alternative splicing networks in primary beta-cells
- PMID: 19934004
- PMCID: PMC2809955
- DOI: 10.2337/db09-1159
Cytokines interleukin-1beta and tumor necrosis factor-alpha regulate different transcriptional and alternative splicing networks in primary beta-cells
Abstract
Objective: Cytokines contribute to pancreatic beta-cell death in type 1 diabetes. This effect is mediated by complex gene networks that remain to be characterized. We presently utilized array analysis to define the global expression pattern of genes, including spliced variants, modified by the cytokines interleukin (IL)-1beta + interferon (IFN)-gamma and tumor necrosis factor (TNF)-alpha + IFN-gamma in primary rat beta-cells.
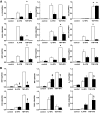
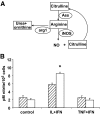
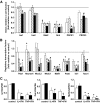
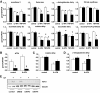
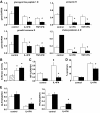
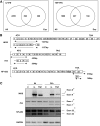
Research design and methods: Fluorescence-activated cell sorter-purified rat beta-cells were exposed to IL-1beta + IFN-gamma or TNF-alpha + IFN-gamma for 6 or 24 h, and global gene expression was analyzed by microarray. Key results were confirmed by RT-PCR, and small-interfering RNAs were used to investigate the mechanistic role of novel and relevant transcription factors identified by pathway analysis. RESULTS Nearly 16,000 transcripts were detected as present in beta-cells, with temporal differences in the number of genes modulated by IL-1beta + IFNgamma or TNF-alpha + IFN-gamma. These cytokine combinations induced differential expression of inflammatory response genes, which is related to differential induction of IFN regulatory factor-7. Both treatments decreased the expression of genes involved in the maintenance of beta-cell phenotype and growth/regeneration. Cytokines induced hypoxia-inducible factor-alpha, which in this context has a proapoptotic role. Cytokines also modified the expression of >20 genes involved in RNA splicing, and exon array analysis showed cytokine-induced changes in alternative splicing of >50% of the cytokine-modified genes.
Conclusions: The present study doubles the number of known genes expressed in primary beta-cells, modified or not by cytokines, and indicates the biological role for several novel cytokine-modified pathways in beta-cells. It also shows that cytokines modify alternative splicing in beta-cells, opening a new avenue of research for the field.
Figures

Comment in
-
Cytokine-induced dicing and splicing in the beta-cell and the immune response in type 1 diabetes.Diabetes. 2010 Feb;59(2):335-6. doi: 10.2337/db09-1767. Diabetes. 2010. PMID: 20103713 Free PMC article. No abstract available.
References
-
- Eizirik DL, Mandrup-Poulsen T: A choice of death-the signal-transduction of immune-mediated β-cell apoptosis. Diabetologia 2001;44:2115–2133 - PubMed
-
- Kaminitz A, Stein J, Yaniv I, Askenasy N: The vicious cycle of apoptotic β-cell death in type 1 diabetes. Immunol Cell Biol 2007;85:582–589 - PubMed
-
- Uno S, Imagawa A, Okita K, Sayama K, Moriwaki M, Iwahashi H, Yamagata K, Tamura S, Matsuzawa Y, Hanafusa T, Miyagawa J, Shimomura I: Macrophages and dendritic cells infiltrating islets with or without β-cells produce tumour necrosis factor-α in patients with recent-onset type 1 diabetes. Diabetologia 2007;50:596–601 - PubMed
-
- Pickersgill LM, Mandrup-Poulsen TR: The anti-interleukin-1 in type 1 diabetes action trial-background and rationale. Diabetes Metab Res Rev 2009;25:321–324 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

