N7-Methylguanine at position 46 (m7G46) in tRNA from Thermus thermophilus is required for cell viability at high temperatures through a tRNA modification network
- PMID: 19934251
- PMCID: PMC2817472
- DOI: 10.1093/nar/gkp1059
N7-Methylguanine at position 46 (m7G46) in tRNA from Thermus thermophilus is required for cell viability at high temperatures through a tRNA modification network
Abstract
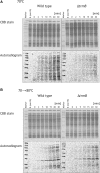
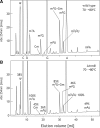
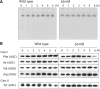
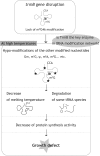
N(7)-methylguanine at position 46 (m(7)G46) in tRNA is produced by tRNA (m(7)G46) methyltransferase (TrmB). To clarify the role of this modification, we made a trmB gene disruptant (DeltatrmB) of Thermus thermophilus, an extreme thermophilic eubacterium. The absence of TrmB activity in cell extract from the DeltatrmB strain and the lack of the m(7)G46 modification in tRNA(Phe) were confirmed by enzyme assay, nucleoside analysis and RNA sequencing. When the DeltatrmB strain was cultured at high temperatures, several modified nucleotides in tRNA were hypo-modified in addition to the lack of the m(7)G46 modification. Assays with tRNA modification enzymes revealed hypo-modifications of Gm18 and m(1)G37, suggesting that the m(7)G46 positively affects their formations. Although the lack of the m(7)G46 modification and the hypo-modifications do not affect the Phe charging activity of tRNA(Phe), they cause a decrease in melting temperature of class I tRNA and degradation of tRNA(Phe) and tRNA(Ile). (35)S-Met incorporation into proteins revealed that protein synthesis in DeltatrmB cells is depressed above 70 degrees C. At 80 degrees C, the DeltatrmB strain exhibits a severe growth defect. Thus, the m(7)G46 modification is required for cell viability at high temperatures via a tRNA modification network, in which the m(7)G46 modification supports introduction of other modifications.
Figures




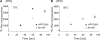
Similar articles
-
Pseudouridine at position 55 in tRNA controls the contents of other modified nucleotides for low-temperature adaptation in the extreme-thermophilic eubacterium Thermus thermophilus.Nucleic Acids Res. 2011 Mar;39(6):2304-18. doi: 10.1093/nar/gkq1180. Epub 2010 Nov 18. Nucleic Acids Res. 2011. PMID: 21097467 Free PMC article.
-
Effects of polyamines from Thermus thermophilus, an extreme-thermophilic eubacterium, on tRNA methylation by tRNA (Gm18) methyltransferase (TrmH).J Biochem. 2016 May;159(5):509-17. doi: 10.1093/jb/mvv130. Epub 2015 Dec 31. J Biochem. 2016. PMID: 26721905
-
Amount changes of tRNA modification enzymes in Thermus thermophilus HB8 cells according to culture temperatures.Nucleic Acids Symp Ser (Oxf). 2006;(50):247-8. doi: 10.1093/nass/nrl123. Nucleic Acids Symp Ser (Oxf). 2006. PMID: 17150910
-
Trm5 and TrmD: Two Enzymes from Distinct Origins Catalyze the Identical tRNA Modification, m¹G37.Biomolecules. 2017 Mar 21;7(1):32. doi: 10.3390/biom7010032. Biomolecules. 2017. PMID: 28335556 Free PMC article. Review.
-
tRNA-m1G methyltransferase interactions: touching bases with structure.Biochimie. 1995;77(1-2):62-5. doi: 10.1016/0300-9084(96)88105-3. Biochimie. 1995. PMID: 7599277 Review.
Cited by
-
RNA modifications in aging-associated cardiovascular diseases.Aging (Albany NY). 2022 Sep 29;14(19):8110-8136. doi: 10.18632/aging.204311. Epub 2022 Sep 29. Aging (Albany NY). 2022. PMID: 36178367 Free PMC article. Review.
-
The occurrence order and cross-talk of different tRNA modifications.Sci China Life Sci. 2021 Sep;64(9):1423-1436. doi: 10.1007/s11427-020-1906-4. Epub 2021 Apr 19. Sci China Life Sci. 2021. PMID: 33881742 Review.
-
Multisite-specific archaeosine tRNA-guanine transglycosylase (ArcTGT) from Thermoplasma acidophilum, a thermo-acidophilic archaeon.Nucleic Acids Res. 2016 Feb 29;44(4):1894-908. doi: 10.1093/nar/gkv1522. Epub 2015 Dec 31. Nucleic Acids Res. 2016. PMID: 26721388 Free PMC article.
-
tRNAVal allows four-way decoding with unmodified uridine at the wobble position in Lactobacillus casei.RNA. 2024 Nov 18;30(12):1608-1619. doi: 10.1261/rna.080155.124. RNA. 2024. PMID: 39255994
-
tRNA pseudouridine synthase D (TruD) from Thermus thermophilus modifies U13 in tRNAAsp, tRNAGlu, and tRNAGln and U35 in tRNATyr.RNA. 2025 May 16;31(6):850-867. doi: 10.1261/rna.080405.125. RNA. 2025. PMID: 40138658
References
-
- Robertus JD, Ladner JE, Finch JT, Rhodes D, Brown RS, Clark BF, Klug A. Structure of yeast phenylalanine tRNA at 3 A resolution. Nature. 1974;250:546–551. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

