SIRT1 deacetylates APE1 and regulates cellular base excision repair
- PMID: 19934257
- PMCID: PMC2817463
- DOI: 10.1093/nar/gkp1039
SIRT1 deacetylates APE1 and regulates cellular base excision repair
Abstract
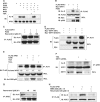
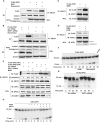
Apurinic/apyrimidinic endonuclease-1 (APE1) is an essential enzyme in the base excision repair (BER) pathway. Here, we show that APE1 is a target of the SIRTUIN1 (SIRT1) protein deacetylase. SIRT1 associates with APE1, and this association is increased with genotoxic stress. SIRT1 deacetylates APE1 in vitro and in vivo targeting lysines 6 and 7. Genotoxic insults stimulate lysine acetylation of APE1 which is antagonized by transcriptional upregulation of SIRT1. Knockdown of SIRT1 increases cellular abasic DNA content, sensitizing cells to death induced by genotoxic stress, and this vulnerability is rescued by overexpression of APE1. Activation of SIRT1 with resveratrol promotes binding of APE1 to the BER protein X-ray cross-complementing-1 (XRCC1), while inhibition of SIRT1 with nicotinamide (NAM) decreases this interaction. Genotoxic insult also increases binding of APE1 to XRCC1, and this increase is suppressed by NAM or knockdown of SIRT1. Finally, resveratrol increases APE activity in XRCC1-associated protein complexes, while NAM or knockdown of SIRT1 suppresses this DNA repair activity. These findings identify APE1 as a novel protein target of SIRT1, and suggest that SIRT1 plays a vital role in maintaining genomic integrity through regulation of the BER pathway.
Figures

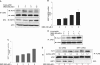



Similar articles
-
Nucleolar accumulation of APE1 depends on charged lysine residues that undergo acetylation upon genotoxic stress and modulate its BER activity in cells.Mol Biol Cell. 2012 Oct;23(20):4079-96. doi: 10.1091/mbc.E12-04-0299. Epub 2012 Aug 23. Mol Biol Cell. 2012. PMID: 22918947 Free PMC article.
-
Elevated level of acetylation of APE1 in tumor cells modulates DNA damage repair.Oncotarget. 2016 Nov 15;7(46):75197-75209. doi: 10.18632/oncotarget.12113. Oncotarget. 2016. PMID: 27655688 Free PMC article.
-
Modulation of the Apurinic/Apyrimidinic Endonuclease Activity of Human APE1 and of Its Natural Polymorphic Variants by Base Excision Repair Proteins.Int J Mol Sci. 2020 Sep 28;21(19):7147. doi: 10.3390/ijms21197147. Int J Mol Sci. 2020. PMID: 32998246 Free PMC article.
-
Functions of the major abasic endonuclease (APE1) in cell viability and genotoxin resistance.Mutagenesis. 2020 Feb 13;35(1):27-38. doi: 10.1093/mutage/gez046. Mutagenesis. 2020. PMID: 31816044 Free PMC article. Review.
-
Inhibitors of nuclease and redox activity of apurinic/apyrimidinic endonuclease 1/redox effector factor 1 (APE1/Ref-1).Bioorg Med Chem. 2017 May 1;25(9):2531-2544. doi: 10.1016/j.bmc.2017.01.028. Epub 2017 Jan 21. Bioorg Med Chem. 2017. PMID: 28161249 Review.
Cited by
-
Post-Translational Modifications by Lipid Metabolites during the DNA Damage Response and Their Role in Cancer.Biomolecules. 2022 Nov 8;12(11):1655. doi: 10.3390/biom12111655. Biomolecules. 2022. PMID: 36359005 Free PMC article. Review.
-
Critical lysine residues within the overlooked N-terminal domain of human APE1 regulate its biological functions.Nucleic Acids Res. 2010 Dec;38(22):8239-56. doi: 10.1093/nar/gkq691. Epub 2010 Aug 10. Nucleic Acids Res. 2010. PMID: 20699270 Free PMC article.
-
Endogenous oxidized DNA bases and APE1 regulate the formation of G-quadruplex structures in the genome.Proc Natl Acad Sci U S A. 2020 May 26;117(21):11409-11420. doi: 10.1073/pnas.1912355117. Epub 2020 May 13. Proc Natl Acad Sci U S A. 2020. PMID: 32404420 Free PMC article.
-
Molecular Mechanisms Regulating the DNA Repair Protein APE1: A Focus on Its Flexible N-Terminal Tail Domain.Int J Mol Sci. 2021 Jun 11;22(12):6308. doi: 10.3390/ijms22126308. Int J Mol Sci. 2021. PMID: 34208390 Free PMC article. Review.
-
Reactive oxygen species, cellular redox systems, and apoptosis.Free Radic Biol Med. 2010 Mar 15;48(6):749-62. doi: 10.1016/j.freeradbiomed.2009.12.022. Epub 2010 Jan 4. Free Radic Biol Med. 2010. PMID: 20045723 Free PMC article. Review.
References
-
- Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 1994;63:915–948. - PubMed
-
- Izumi T, Wiederhold LR, Roy G, Roy R, Jaiswal A, Bhakat KK, Mitra S, Hazra TK. Mammalian DNA base excision repair proteins: their interactions and role in repair of oxidative DNA damage. Toxicology. 2003;193:43–65. - PubMed
-
- Mitra S, Izumi T, Boldogh I, Bhakat KK, Hill JW, Hazra TK. Choreography of oxidative damage repair in mammalian genomes. Free Radic. Biol. Med. 2002;33:15–28. - PubMed
-
- Tell G, Damante G, Caldwell D, Kelley MR. The intracellular localization of APE1/Ref-1: more than a passive phenomenon? Antioxid. Redox Signal. 2005;7:367–384. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

