The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis
- PMID: 19934380
- PMCID: PMC2798333
- DOI: 10.1105/tpc.108.064980
The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis
Abstract
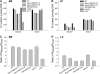
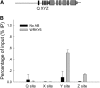
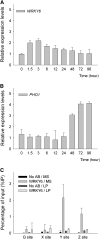
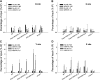
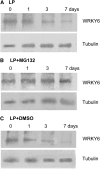
Arabidopsis thaliana WRKY family comprises 74 members and some of them are involved in plant responses to biotic and abiotic stresses. This study demonstrated that WRKY6 is involved in Arabidopsis responses to low-Pi stress through regulating PHOSPHATE1 (PHO1) expression. WRKY6 overexpression lines, similar to the pho1 mutant, were more sensitive to low Pi stress and had lower Pi contents in shoots compared with wild-type seedlings and the wrky6-1 mutant. Immunoprecipitation assays demonstrated that WRKY6 can bind to two W-boxes of the PHO1 promoter. RNA gel blot and beta-glucuronidase activity assays showed that PHO1 expression was repressed in WRKY6-overexpressing lines and enhanced in the wrky6-1 mutant. Low Pi treatment reduced WRKY6 binding to the PHO1 promoter, which indicates that PHO1 regulation by WRKY6 is Pi dependent and that low Pi treatment may release inhibition of PHO1 expression. Protein gel blot analysis showed that the decrease in WRKY6 protein induced by low Pi treatment was inhibited by a 26S proteosome inhibitor, MG132, suggesting that low Pi-induced release of PHO1 repression may result from 26S proteosome-mediated proteolysis. In addition, WRKY42 also showed binding to W-boxes of the PHO1 promoter and repressed PHO1 expression. Our results demonstrate that WRKY6 and WRKY42 are involved in Arabidopsis responses to low Pi stress by regulation of PHO1 expression.
Figures



References
-
- Ames, B.N. (1966). Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 8 115–118.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

