Protein kinase C: poised to signal
- PMID: 19934406
- PMCID: PMC2838521
- DOI: 10.1152/ajpendo.00477.2009
Protein kinase C: poised to signal
Abstract
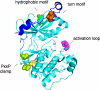
Nestled at the tip of a branch of the kinome, protein kinase C (PKC) family members are poised to transduce signals emanating from the cell surface. Cell membranes provide the platform for PKC function, supporting the maturation of PKC through phosphorylation, its allosteric activation by binding specific lipids, and, ultimately, promoting the downregulation of the enzyme. These regulatory mechanisms precisely control the level of signaling-competent PKC in the cell. Disruption of this regulation results in pathophysiological states, most notably cancer, where PKC levels are often grossly altered. This review introduces the PKC family and then focuses on recent advances in understanding the cellular regulation of its diacylglycerol-regulated members.
Figures


References
-
- Akamine P, Madhusudan Wu J, Xuong NH, Ten Eyck LF, Taylor SS. Dynamic features of cAMP-dependent protein kinase revealed by apoenzyme crystal structure. J Mol Biol 327: 159–171, 2003 - PubMed
-
- Aziz MH, Manoharan HT, Church DR, Dreckschmidt NE, Zhong W, Oberley TD, Wilding G, Verma AK. Protein kinase Cepsilon interacts with signal transducers and activators of transcription 3 (Stat3), phosphorylates Stat3Ser727, and regulates its constitutive activation in prostate cancer. Cancer Res 67: 8828–8838, 2007 - PubMed
-
- Balendran A, Hare GR, Kieloch A, Williams MR, Alessi DR. Further evidence that 3-phosphoinositide-dependent protein kinase-1 (PDK1) is required for the stability and phosphorylation of protein kinase C (PKC) isoforms. FEBS Lett 484: 217–223, 2000 - PubMed
-
- Bartlett PJ, Young KW, Nahorski SR, Challiss RA. Single cell analysis and temporal profiling of agonist-mediated inositol 1,4,5-trisphosphate, Ca2+, diacylglycerol, and protein kinase C signaling using fluorescent biosensors. J Biol Chem 280: 21837–21846, 2005 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

