Exploring protein fitness landscapes by directed evolution
- PMID: 19935669
- PMCID: PMC2997618
- DOI: 10.1038/nrm2805
Exploring protein fitness landscapes by directed evolution
Abstract
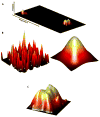
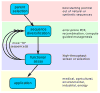
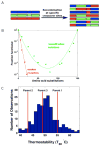
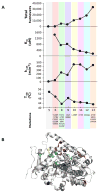
Directed evolution circumvents our profound ignorance of how a protein's sequence encodes its function by using iterative rounds of random mutation and artificial selection to discover new and useful proteins. Proteins can be tuned to adapt to new functions or environments by simple adaptive walks involving small numbers of mutations. Directed evolution studies have shown how rapidly some proteins can evolve under strong selection pressures and, because the entire 'fossil record' of evolutionary intermediates is available for detailed study, they have provided new insight into the relationship between sequence and function. Directed evolution has also shown how mutations that are functionally neutral can set the stage for further adaptation.
Figures
References
-
- Chen K, Arnold FH. Tuning the activity of an enzyme for unusual environments: sequential random mutagenesis of subtilisin E for catalysis in dimethylformamide. Proc Natl Acad Sci USA. 1993;90:5618–5622. - PMC - PubMed
-
The first demonstration of directed evolution by successive rounds of mutagenesis and screening, a strategy now widely used to engineer enzymes.
-
- Reetz MT. Combinatorial and evolution-based methods in the creation of enantioselective catalysts. Angew Chem Int Ed. 2001;40:284–310. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

