Caenorhabditis elegans HIM-18/SLX-4 interacts with SLX-1 and XPF-1 and maintains genomic integrity in the germline by processing recombination intermediates
- PMID: 19936019
- PMCID: PMC2770170
- DOI: 10.1371/journal.pgen.1000735
Caenorhabditis elegans HIM-18/SLX-4 interacts with SLX-1 and XPF-1 and maintains genomic integrity in the germline by processing recombination intermediates
Abstract
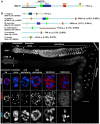
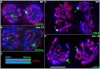
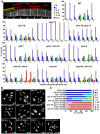
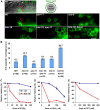
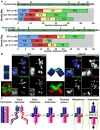
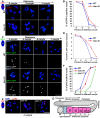
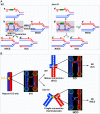
Homologous recombination (HR) is essential for the repair of blocked or collapsed replication forks and for the production of crossovers between homologs that promote accurate meiotic chromosome segregation. Here, we identify HIM-18, an ortholog of MUS312/Slx4, as a critical player required in vivo for processing late HR intermediates in Caenorhabditis elegans. DNA damage sensitivity and an accumulation of HR intermediates (RAD-51 foci) during premeiotic entry suggest that HIM-18 is required for HR-mediated repair at stalled replication forks. A reduction in crossover recombination frequencies-accompanied by an increase in HR intermediates during meiosis, germ cell apoptosis, unstable bivalent attachments, and subsequent chromosome nondisjunction-support a role for HIM-18 in converting HR intermediates into crossover products. Such a role is suggested by physical interaction of HIM-18 with the nucleases SLX-1 and XPF-1 and by the synthetic lethality of him-18 with him-6, the C. elegans BLM homolog. We propose that HIM-18 facilitates processing of HR intermediates resulting from replication fork collapse and programmed meiotic DSBs in the C. elegans germline.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. - PubMed
-
- Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. - PubMed
-
- Lilley DM, White MF. The junction-resolving enzymes. Nat Rev Mol Cell Biol. 2001;2:433–443. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

