Fine-scale phylogenetic discordance across the house mouse genome
- PMID: 19936022
- PMCID: PMC2770633
- DOI: 10.1371/journal.pgen.1000729
Fine-scale phylogenetic discordance across the house mouse genome
Abstract
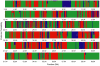
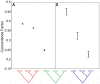
Population genetic theory predicts discordance in the true phylogeny of different genomic regions when studying recently diverged species. Despite this expectation, genome-wide discordance in young species groups has rarely been statistically quantified. The house mouse subspecies group provides a model system for examining phylogenetic discordance. House mouse subspecies are recently derived, suggesting that even if there has been a simple tree-like population history, gene trees could disagree with the population history due to incomplete lineage sorting. Subspecies of house mice also hybridize in nature, raising the possibility that recent introgression might lead to additional phylogenetic discordance. Single-locus approaches have revealed support for conflicting topologies, resulting in a subspecies tree often summarized as a polytomy. To analyze phylogenetic histories on a genomic scale, we applied a recently developed method, Bayesian concordance analysis, to dense SNP data from three closely related subspecies of house mice: Mus musculus musculus, M. m. castaneus, and M. m. domesticus. We documented substantial variation in phylogenetic history across the genome. Although each of the three possible topologies was strongly supported by a large number of loci, there was statistical evidence for a primary phylogenetic history in which M. m. musculus and M. m. castaneus are sister subspecies. These results underscore the importance of measuring phylogenetic discordance in other recently diverged groups using methods such as Bayesian concordance analysis, which are designed for this purpose.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Rokas A, Williams BL, King N, Carroll SB. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature. 2003;425:798–804. - PubMed
-
- Hackett SJ, Kimball RT, Reddy S, Bowie RC, Braun EL, et al. A phylogenomic study of birds reveals their evolutionary history. Science. 2008;320:1763–1768. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

