Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome
- PMID: 19936065
- PMCID: PMC2774510
- DOI: 10.1371/journal.pgen.1000732
Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome
Abstract
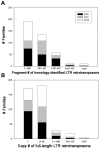
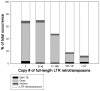
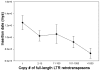
Recent comprehensive sequence analysis of the maize genome now permits detailed discovery and description of all transposable elements (TEs) in this complex nuclear environment. Reiteratively optimized structural and homology criteria were used in the computer-assisted search for retroelements, TEs that transpose by reverse transcription of an RNA intermediate, with the final results verified by manual inspection. Retroelements were found to occupy the majority (>75%) of the nuclear genome in maize inbred B73. Unprecedented genetic diversity was discovered in the long terminal repeat (LTR) retrotransposon class of retroelements, with >400 families (>350 newly discovered) contributing >31,000 intact elements. The two other classes of retroelements, SINEs (four families) and LINEs (at least 30 families), were observed to contribute 1,991 and approximately 35,000 copies, respectively, or a combined approximately 1% of the B73 nuclear genome. With regard to fully intact elements, median copy numbers for all retroelement families in maize was 2 because >250 LTR retrotransposon families contained only one or two intact members that could be detected in the B73 draft sequence. The majority, perhaps all, of the investigated retroelement families exhibited non-random dispersal across the maize genome, with LINEs, SINEs, and many low-copy-number LTR retrotransposons exhibiting a bias for accumulation in gene-rich regions. In contrast, most (but not all) medium- and high-copy-number LTR retrotransposons were found to preferentially accumulate in gene-poor regions like pericentromeric heterochromatin, while a few high-copy-number families exhibited the opposite bias. Regions of the genome with the highest LTR retrotransposon density contained the lowest LTR retrotransposon diversity. These results indicate that the maize genome provides a great number of different niches for the survival and procreation of a great variety of retroelements that have evolved to differentially occupy and exploit this genomic diversity.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- McClintock B. Mutable loci in maize. Year Book: Carnegie Institute of Washington; 1948. pp. 155–169.
-
- Berg DE, Howe MM. Mobile DNA. Washington, DC: American Society for Microbiology; 1989.
-
- Bennetzen JL. Transposable elements, gene creation and genome rearrangement in flowering plants. Curr Opin Genet Dev. 2005;15:621–627. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

