Genomic loci modulating the retinal transcriptome in wound healing
- PMID: 19936100
- PMCID: PMC2759132
Genomic loci modulating the retinal transcriptome in wound healing
Abstract
Purpose: The present study predicts and tests genetic networks that modulate gene expression during the retinal wound-healing response.
Methods: Upstream modulators and target genes were defined using meta-analysis and bioinformatic approaches. Quantitative trait loci (QTLs) for retinal acute phase genes (Vazquez-Chona et al. 2005) were defined using QTL analysis of CNS gene expression (Chesler et al. 2005). Candidate modulators were defined using computational analysis of gene and motif sequences. The effect of candidate genes on wound healing was tested using animal models of gene expression.
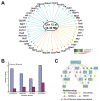
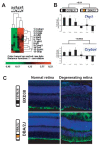
Results: A network of early wound-healing genes is modulated by a locus on chromosome 12. The genetic background of the locus altered the wound-healing response of the retina. The C57BL/6 allele conferred enhanced expression of neuronal marker Thy1 and heat-shock-like crystallins, whereas the DBA/2J allele correlated with greater levels of the classic marker of retinal stress, glial fibrillary acidic protein (GFAP). Id2 and Lpin1 are candidate upstream modulators as they strongly correlated with the segregation of DBA/2J and C57BL/6 alleles, and their dosage levels correlated with the enhanced expression of survival genes (Thy1 and crystallin genes).
Conclusion: We defined a genetic network associated with the retinal acute injury response. Using genetic linkage analysis of natural transcript variation, we identified regulatory loci and can didate modulators that control transcript levels of acute phase genes. Our results support the convergence of gene expression profiling, QTL analysis, and bioinformatics as a rational approach to discover molecular pathways controlling retinal wound healing.
Keywords: CNS degeneration; QTL analysis; genetic networks; microarray; retinal degeneration.
Figures



Similar articles
-
Genetic networks controlling retinal injury.Mol Vis. 2005 Nov 4;11:958-70. Mol Vis. 2005. PMID: 16288200 Free PMC article.
-
Quantitative genetics of age-related retinal degeneration: a second F1 intercross between the A/J and C57BL/6 strains.Mol Vis. 2007 Jan 25;13:79-85. Mol Vis. 2007. PMID: 17277741 Free PMC article.
-
Using gene expression databases for classical trait QTL candidate gene discovery in the BXD recombinant inbred genetic reference population: mouse forebrain weight.BMC Genomics. 2008 Sep 25;9:444. doi: 10.1186/1471-2164-9-444. BMC Genomics. 2008. PMID: 18817551 Free PMC article.
-
Genomic locus modulating corneal thickness in the mouse identifies POU6F2 as a potential risk of developing glaucoma.PLoS Genet. 2018 Jan 25;14(1):e1007145. doi: 10.1371/journal.pgen.1007145. eCollection 2018 Jan. PLoS Genet. 2018. PMID: 29370175 Free PMC article.
-
Use of recombinant inbred strains for studying genetic determinants of responses to alcohol.Alcohol Alcohol Suppl. 1994;2:67-71. Alcohol Alcohol Suppl. 1994. PMID: 8974318 Review.
Cited by
-
Association of high myopia with crystallin beta A4 (CRYBA4) gene polymorphisms in the linkage-identified MYP6 locus.PLoS One. 2012;7(6):e40238. doi: 10.1371/journal.pone.0040238. Epub 2012 Jun 29. PLoS One. 2012. PMID: 22792142 Free PMC article.
-
Robust classification of wound healing stages in both mice and humans for acute and burn wounds based on transcriptomic data.BMC Bioinformatics. 2023 Apr 25;24(1):166. doi: 10.1186/s12859-023-05295-z. BMC Bioinformatics. 2023. PMID: 37098473 Free PMC article.
-
Using BXD mouse strains in vision research: A systems genetics approach.Mol Vis. 2020 Mar 6;26:173-187. eCollection 2020. Mol Vis. 2020. PMID: 32180682 Free PMC article. Review.
-
Networks modulating the retinal response to injury: insights from microarrays, expression genetics, and bioinformatics.Adv Exp Med Biol. 2012;723:649-56. doi: 10.1007/978-1-4614-0631-0_82. Adv Exp Med Biol. 2012. PMID: 22183389 Free PMC article. Review. No abstract available.
-
A practical approach to optic nerve crush in the mouse.Mol Vis. 2012;18:2147-52. Epub 2012 Jul 27. Mol Vis. 2012. PMID: 22876142 Free PMC article.
References
-
- Ahmed F, Brown KM, Stephan DA, Morrison JC, Johnson EC, Tomarev SI. Microarray analysis of changes in mRNA levels in the rat retina after experimental elevation of intraocular pressure. Invest Ophthalmol Vis Sci. 2004;45:1247–58. - PubMed
-
- Aitman TJ, Glazier AM, Wallace CA, Cooper LD, Norsworthy PJ, Wahid FN, Al-Majali KM, Trembling PM, Mann CJ, Shoulders CC, Graf D, St Lezin E, Kurtz TW, Kren V, Pravenec M, Ibrahimi A, Abumrad NA, Stanton LW, Scott J. Identification of Cd36 (Fat) as an insulin-resistance gene causing defective fatty acid and glucose metabolism in hypertensive rats. Nat Genet. 1999;21:76–83. - PubMed
-
- Aronica E, Vandeputte DA, Van Vliet EA, Lopes DA Silva FH, Troost D, Gorter JA. Expression of Id proteins increases in astrocytes in the hippocampus of epileptic rats. Neuroreport. 2001;12:2461–5. - PubMed
-
- Berge KE, Tian H, Graf GA, Yu L, Grishin NV, Schultz J, Kwiterovich P, Shan B, Barnes R, Hobbs HH. Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent ABC transporters. Science. 2000;290:1771–5. - PubMed
-
- Broman KW. Mapping expression in randomized rodent genomes. Nat Genet. 2005;37:209–10. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
