beta-Strand interactions at the domain interface critical for the stability of human lens gammaD-crystallin
- PMID: 19937657
- PMCID: PMC2817848
- DOI: 10.1002/pro.296
beta-Strand interactions at the domain interface critical for the stability of human lens gammaD-crystallin
Abstract
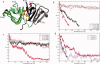
Human age-onset cataracts are believed to be caused by the aggregation of partially unfolded or covalently damaged lens crystallin proteins; however, the exact molecular mechanism remains largely unknown. We have used microseconds of molecular dynamics simulations with explicit solvent to investigate the unfolding process of human lens gammaD-crystallin protein and its isolated domains. A partially unfolded folding intermediate of gammaD-crystallin is detected in simulations with its C-terminal domain (C-td) folded and N-terminal domain (N-td) unstructured, in excellent agreement with biochemical experiments. Our simulations strongly indicate that the stability and the folding mechanism of the N-td are regulated by the interdomain interactions, consistent with experimental observations. A hydrophobic folding core was identified within the C-td that is comprised of a and b strands from the Greek key motif 4, the one near the domain interface. Detailed analyses reveal a surprising non-native surface salt-bridge between Glu135 and Arg142 located at the end of the ab folded hairpin turn playing a critical role in stabilizing the folding core. On the other hand, an in silico single E135A substitution that disrupts this non-native Glu135-Arg142 salt-bridge causes significant destabilization to the folding core of the isolated C-td, which, in turn, induces unfolding of the N-td interface. These findings indicate that certain highly conserved charged residues, that is, Glu135 and Arg142, of gammaD-crystallin are crucial for stabilizing its hydrophobic domain interface in native conformation, and disruption of charges on the gammaD-crystallin surface might lead to unfolding and subsequent aggregation.
Figures
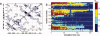


Similar articles
-
Contributions of aromatic pairs to the folding and stability of long-lived human γD-crystallin.Protein Sci. 2011 Mar;20(3):513-28. doi: 10.1002/pro.583. Protein Sci. 2011. PMID: 21432932 Free PMC article.
-
Interdomain side-chain interactions in human gammaD crystallin influencing folding and stability.Protein Sci. 2005 Aug;14(8):2030-43. doi: 10.1110/ps.051460505. Protein Sci. 2005. PMID: 16046626 Free PMC article.
-
Dissecting the contributions of β-hairpin tyrosine pairs to the folding and stability of long-lived human γD-crystallins.Nanoscale. 2014;6(3):1797-807. doi: 10.1039/c3nr03782g. Nanoscale. 2014. PMID: 24352614 Free PMC article.
-
Aggregation of γ-crystallins associated with human cataracts via domain swapping at the C-terminal β-strands.Proc Natl Acad Sci U S A. 2011 Jun 28;108(26):10514-9. doi: 10.1073/pnas.1019152108. Epub 2011 Jun 13. Proc Natl Acad Sci U S A. 2011. PMID: 21670251 Free PMC article.
-
The βγ-crystallins: native state stability and pathways to aggregation.Prog Biophys Mol Biol. 2014 Jul;115(1):32-41. doi: 10.1016/j.pbiomolbio.2014.05.002. Epub 2014 May 14. Prog Biophys Mol Biol. 2014. PMID: 24835736 Free PMC article. Review.
Cited by
-
Cataract-causing defect of a mutant γ-crystallin proceeds through an aggregation pathway which bypasses recognition by the α-crystallin chaperone.PLoS One. 2012;7(5):e37256. doi: 10.1371/journal.pone.0037256. Epub 2012 May 24. PLoS One. 2012. PMID: 22655036 Free PMC article.
-
Evolutionary remodeling of βγ-crystallins for domain stability at cost of Ca2+ binding.J Biol Chem. 2011 Dec 23;286(51):43891-43901. doi: 10.1074/jbc.M111.247890. Epub 2011 Sep 26. J Biol Chem. 2011. PMID: 21949186 Free PMC article.
-
The Effect of Attractive Interactions and Macromolecular Crowding on Crystallins Association.PLoS One. 2016 Mar 8;11(3):e0151159. doi: 10.1371/journal.pone.0151159. eCollection 2016. PLoS One. 2016. PMID: 26954357 Free PMC article.
-
Contributions of aromatic pairs to the folding and stability of long-lived human γD-crystallin.Protein Sci. 2011 Mar;20(3):513-28. doi: 10.1002/pro.583. Protein Sci. 2011. PMID: 21432932 Free PMC article.
-
UV-radiation induced disruption of dry-cavities in human γD-crystallin results in decreased stability and faster unfolding.Sci Rep. 2013;3:1560. doi: 10.1038/srep01560. Sci Rep. 2013. PMID: 23532089 Free PMC article.
References
-
- Pandey SK, Apple DJ, Werner L, Maloof AJ, Milverton EJ. Posterior capsule opacification: a review of the aetiopathogenesis, experimental and clinical studies and factors for prevention. Indian J Ophthalmol. 2004;52:99–112. - PubMed
-
- Benedek GB. Cataract as a protein condensation disease: the Proctor Lecture. Invest Ophthalmol Vis Sci. 1997;38:1911–1921. - PubMed
-
- Aarts HJM, Lubsen NH, Schoenmakers JGG. Crystallin gene-expression during rat lens development. Eur J Biochem. 1989;183:31–36. - PubMed
-
- Lampi KJ, Shih M, Ueda Y, Shearer TR, David LL. Lens proteomics: analysis of rat crystallin sequences and two-dimensional electrophoresis map. Invest Ophthalmol Visual Sci. 2002;43:216–224. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

