High diversity of hepatitis C viral quasispecies is associated with early virological response in patients undergoing antiviral therapy
- PMID: 19937690
- PMCID: PMC2911951
- DOI: 10.1002/hep.23290
High diversity of hepatitis C viral quasispecies is associated with early virological response in patients undergoing antiviral therapy
Abstract
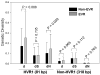
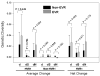
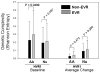
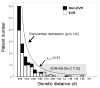
Differential response patterns to optimal antiviral therapy, peginterferon alpha plus ribavirin, are well documented in patients with chronic hepatitis C virus (HCV) infection. Among many factors that may affect therapeutic efficiency, HCV quasispecies (QS) characteristics have been a major focus of previous studies, yielding conflicting results. To obtain a comprehensive understanding of the role of HCV QS in antiviral therapy, we performed the largest-ever HCV QS analysis in 153 patients infected with HCV genotype 1 strains. A total of 4,314 viral clones spanning hypervarible region 1 were produced from these patients during the first 12 weeks of therapy, followed by detailed genetic analyses. Our data show an exponential distribution pattern of intrapatient QS diversity in this study population in which most patients (63%) had small QS diversity with genetic distance (d) less than 0.2. The group of patients with genetic distance located in the decay region (d>0.53) had a significantly higher early virologic response (EVR) rate (89.5%), which contributed substantially to the overall association between EVR and increased baseline QS diversity. In addition, EVR was linked to a clustered evolutionary pattern in terms of QS dynamic changes.
Conclusion: EVR is associated with elevated HCV QS diversity and complexity, especially in patients with significantly higher HCV genetic heterogeneity.
Figures

Similar articles
-
Comparison of amplification enzymes for Hepatitis C Virus quasispecies analysis.Virol J. 2005 Apr 22;2:41. doi: 10.1186/1743-422X-2-41. Virol J. 2005. PMID: 15847697 Free PMC article.
-
Quasispecies of hepatitis C virus genotype 1 and treatment outcome with peginterferon and ribavirin.Infect Genet Evol. 2009 Jul;9(4):689-98. doi: 10.1016/j.meegid.2008.11.001. Epub 2008 Nov 21. Infect Genet Evol. 2009. PMID: 19063998
-
Quasispecies as predictive factor of rapid, early and sustained virological responses in chronic hepatitis C, genotype 1, treated with peginterferon-ribavirin.J Clin Virol. 2008 Apr;41(4):264-9. doi: 10.1016/j.jcv.2007.11.023. Epub 2008 Jan 24. J Clin Virol. 2008. PMID: 18221912
-
Financial impact of two different ways of evaluating early virological response to peginterferon-alpha-2b plus ribavirin therapy in treatment-naive patients with chronic hepatitis C virus genotype 1.Pharmacoeconomics. 2005;23(10):1043-55. doi: 10.2165/00019053-200523100-00007. Pharmacoeconomics. 2005. PMID: 16235977 Review.
-
Hepatitis C virus genetic variability in patients undergoing antiviral therapy.Virus Res. 2007 Aug;127(2):185-94. doi: 10.1016/j.virusres.2007.02.023. Epub 2007 Apr 20. Virus Res. 2007. PMID: 17449128 Review.
Cited by
-
Mutagen resistance and mutation restriction of St. Louis encephalitis virus.J Gen Virol. 2017 Feb;98(2):201-211. doi: 10.1099/jgv.0.000682. J Gen Virol. 2017. PMID: 28284278 Free PMC article.
-
Dynamics of Genotypic Mutations of the Hepatitis B Virus Associated With Long-Term Entecavir Treatment Determined With Ultradeep Pyrosequencing: A Retrospective Observational Study.Medicine (Baltimore). 2016 Jan;95(4):e2614. doi: 10.1097/MD.0000000000002614. Medicine (Baltimore). 2016. PMID: 26825915 Free PMC article.
-
Prospects for personalizing antiviral therapy for hepatitis C virus with pharmacogenetics.Genome Med. 2011 Feb 8;3(2):8. doi: 10.1186/gm222. Genome Med. 2011. PMID: 21345258 Free PMC article.
-
Clinical implications of evolutionary patterns of homologous, full-length hepatitis B virus quasispecies in different hosts after perinatal infection.J Clin Microbiol. 2014 May;52(5):1556-65. doi: 10.1128/JCM.03338-13. Epub 2014 Feb 26. J Clin Microbiol. 2014. PMID: 24574300 Free PMC article.
-
Comparison of the Mechanisms of Drug Resistance among HIV, Hepatitis B, and Hepatitis C.Viruses. 2010 Dec 1;2(12):2696-739. doi: 10.3390/v2122696. Viruses. 2010. PMID: 21243082 Free PMC article.
References
-
- National Institutes of Health. NIH Consensus Development Conference Statement: Management of hepatitis C. Hepatology. 2002;36 (Suppl 1):S3–20. - PubMed
-
- Feld JJ, Hoofnagle JH. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature. 2005;436:967–972. - PubMed
-
- Di Bisceglie AM, Ghalib RH, Hamzeh FM, Rustgi VK. Early virologic response after peginterferon alpha-2a plus ribavirin or peginterferon alpha-2b plus ribavirin treatment in patients with chronic hepatitis C. J Viral Hepatitis. 2007;14:721–729. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
