Investigating solution-phase protein structure and dynamics by hydrogen exchange mass spectrometry
- PMID: 19937720
- PMCID: PMC2844867
- DOI: 10.1002/0471140864.ps1706s58
Investigating solution-phase protein structure and dynamics by hydrogen exchange mass spectrometry
Abstract
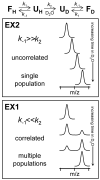
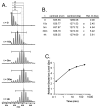
By taking advantage of labeling methods such as hydrogen exchange (HX), many details about protein conformation, dynamics, and interactions can be revealed by mass spectrometry. In this unit, hydrogen exchange theory is discussed as it applies to HX-MS protocols, the practice of HX-MS including data analysis and interpretation is explained in detail, and recent advancements in technology which greatly increase the depth of information gained from the technique are highlighted.
Figures






References
-
- Bai Y, Milne JS, Mayne L, Englander SW. Protein stability parameters measured by hydrogen exchange. Proteins. 1994;20(1):4–14. - PubMed
-
- Bohak Z. Purification and characterization of chicken pepsinogen and chicken pepsin. J Biol Chem. 1969;244(17):4638–4648. - PubMed
-
- Brier S, Engen JR. Hydrogen exchange mass spectrometry: Principles and capabilities. In: Chance M, editor. Mass spectrometry analysis for protein-protein interactions and dynamics. New York: Wiley-Blackwell; 2008. pp. 11–43.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

