Analysis of tandem mass spectra by FTMS for improved large-scale proteomics with superior protein quantification
- PMID: 19938823
- PMCID: PMC2800853
- DOI: 10.1021/ac902005s
Analysis of tandem mass spectra by FTMS for improved large-scale proteomics with superior protein quantification
Abstract
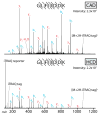
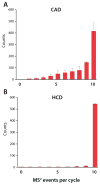
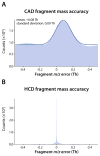
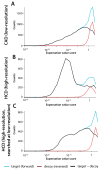
Using a newly developed dual-cell quadrupole linear ion trap-orbitrap hybrid mass spectrometer (dcQLT-orbitrap), we demonstrate the utility of collecting high-resolution tandem mass spectral data for large-scale shotgun proteomics. Multiple nanoLC-MS/MS experiments on both an older generation quadrupole linear ion trap-orbitrap hybrid (QLT-orbitrap) and the dcQLT-orbitrap, using both resonant-excitation CAD and beam-type CAD (HCD), were performed. Resulting from various technological advances (e.g., a stacked ring ion guide AP inlet, a dual cell QLT), the dcQLT-orbitrap exhibited increased duty cycle (approximately 1.5-2 times) and sensitivity for both CAD (ion trap detection) and HCD (orbitrap detection) methods. As compared to the older system, the dcQLT-orbitrap produced significantly more unique peptide identifications for both methods (approximately 30% improvement for CAD and approximately 115% improvement for HCD). The sizable improvement of the HCD method on the dcQLT-orbitrap system outperforms the current standard method of CAD with ion trap detection for large-scale analysis. Finally, we demonstrate that the increased HCD performance translates to a direct and substantial improvement in protein quantitation precision using isobaric tags.
Figures

Similar articles
-
Investigating Acquisition Performance on the Orbitrap Fusion When Using Tandem MS/MS/MS Scanning with Isobaric Tags.J Proteome Res. 2017 May 5;16(5):1839-1846. doi: 10.1021/acs.jproteome.7b00091. Epub 2017 Apr 20. J Proteome Res. 2017. PMID: 28418257
-
Improved precision of iTRAQ and TMT quantification by an axial extraction field in an Orbitrap HCD cell.Anal Chem. 2011 Feb 15;83(4):1469-74. doi: 10.1021/ac102265w. Epub 2011 Jan 28. Anal Chem. 2011. PMID: 21275378 Free PMC article.
-
Ultra high resolution linear ion trap Orbitrap mass spectrometer (Orbitrap Elite) facilitates top down LC MS/MS and versatile peptide fragmentation modes.Mol Cell Proteomics. 2012 Mar;11(3):O111.013698. doi: 10.1074/mcp.O111.013698. Epub 2011 Dec 9. Mol Cell Proteomics. 2012. PMID: 22159718 Free PMC article.
-
Evaluation of the Orbitrap Ascend Tribrid Mass Spectrometer for Shotgun Proteomics.Anal Chem. 2023 Jul 18;95(28):10655-10663. doi: 10.1021/acs.analchem.3c01155. Epub 2023 Jun 30. Anal Chem. 2023. PMID: 37389810 Free PMC article.
-
Screening of synthetic PDE-5 inhibitors and their analogues as adulterants: analytical techniques and challenges.J Pharm Biomed Anal. 2014 Jan;87:176-90. doi: 10.1016/j.jpba.2013.04.037. Epub 2013 May 6. J Pharm Biomed Anal. 2014. PMID: 23721687 Review.
Cited by
-
Amine-reactive neutron-encoded labels for highly plexed proteomic quantitation.Mol Cell Proteomics. 2013 Nov;12(11):3360-9. doi: 10.1074/mcp.M113.032011. Epub 2013 Jul 23. Mol Cell Proteomics. 2013. PMID: 23882030 Free PMC article.
-
Proteomic strategies and challenges in tumor metastasis research.Clin Exp Metastasis. 2010 Aug;27(6):441-51. doi: 10.1007/s10585-010-9339-7. Epub 2010 Jul 7. Clin Exp Metastasis. 2010. PMID: 20607365 Review.
-
Current algorithmic solutions for peptide-based proteomics data generation and identification.Curr Opin Biotechnol. 2013 Feb;24(1):31-8. doi: 10.1016/j.copbio.2012.10.013. Epub 2012 Nov 8. Curr Opin Biotechnol. 2013. PMID: 23142544 Free PMC article. Review.
-
Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics.Mol Cell Proteomics. 2010 Aug;9(8):1634-49. doi: 10.1074/mcp.R000001-MCP201. Epub 2010 Mar 31. Mol Cell Proteomics. 2010. PMID: 20360032 Free PMC article. Review.
-
A proteomics search algorithm specifically designed for high-resolution tandem mass spectra.J Proteome Res. 2013 Mar 1;12(3):1377-86. doi: 10.1021/pr301024c. Epub 2013 Jan 31. J Proteome Res. 2013. PMID: 23323968 Free PMC article.
References
-
- Washburn MP, Wolters D, Yates JR. Nat Biotechnol. 2001;19:242. - PubMed
-
- de Godoy LMF, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M. Nature. 2008;455:1251. - PubMed
-
- Mcluckey SA, Glish GL, Asano KG. Anal Chim Acta. 1989;225:25.
-
- Vanberkel GJ, Glish GL, Mcluckey SA. Analytical Chemistry. 1990;62:1284.
-
- Vanberkel GJ, Mcluckey SA, Glish GL. Analytical Chemistry. 1991;63:1098. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

