Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies
- PMID: 19939925
- PMCID: PMC2812321
- DOI: 10.1128/JVI.02108-09
Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies
Abstract
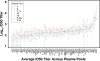
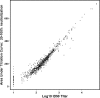
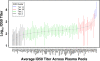
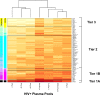
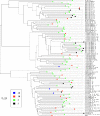
The restricted neutralization breadth of vaccine-elicited antibodies is a major limitation of current human immunodeficiency virus-1 (HIV-1) candidate vaccines. In order to permit the efficient identification of vaccines with enhanced capacity for eliciting cross-reactive neutralizing antibodies (NAbs) and to assess the overall breadth and potency of vaccine-elicited NAb reactivity, we assembled a panel of 109 molecularly cloned HIV-1 Env pseudoviruses representing a broad range of genetic and geographic diversity. Viral isolates from all major circulating genetic subtypes were included, as were viruses derived shortly after transmission and during the early and chronic stages of infection. We assembled a panel of genetically diverse HIV-1-positive (HIV-1(+)) plasma pools to assess the neutralization sensitivities of the entire virus panel. When the viruses were rank ordered according to the average sensitivity to neutralization by the HIV-1(+) plasmas, a continuum of average sensitivity was observed. Clustering analysis of the patterns of sensitivity defined four subgroups of viruses: those having very high (tier 1A), above-average (tier 1B), moderate (tier 2), or low (tier 3) sensitivity to antibody-mediated neutralization. We also investigated potential associations between characteristics of the viral isolates (clade, stage of infection, and source of virus) and sensitivity to NAb. In particular, higher levels of NAb activity were observed when the virus and plasma pool were matched in clade. These data provide the first systematic assessment of the overall neutralization sensitivities of a genetically and geographically diverse panel of circulating HIV-1 strains. These reference viruses can facilitate the systematic characterization of NAb responses elicited by candidate vaccine immunogens.
Figures



References
-
- Baba, T. W., V. Liska, R. Hofmann-Lehmann, J. Vlasak, W. Xu, S. Ayehunie, L. A. Cavacini, M. R. Posner, H. Katinger, G. Stiegler, B. J. Bernacky, T. A. Rizvi, R. Schmidt, L. R. Hill, M. E. Keeling, Y. Lu, J. E. Wright, T. C. Chou, and R. M. Ruprecht. 2000. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat. Med. 6:200-206. - PubMed
-
- Bhattacharya, T., M. Daniels, D. Heckerman, B. Foley, N. Frahm, C. Kadie, J. Carlson, K. Yusim, B. McMahon, B. Gaschen, S. Mallal, J. I. Mullins, D. C. Nickle, J. Herbeck, C. Rousseau, G. H. Learn, T. Miura, C. Brander, B. Walker, and B. Korber. 2007. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science 315:1583-1586. - PubMed
-
- Binley, J. M., E. A. Lybarger, E. T. Crooks, M. S. Seaman, E. Gray, K. L. Davis, J. M. Decker, D. Wycuff, L. Harris, N. Hawkins, B. Wood, C. Nathe, D. Richman, G. D. Tomaras, F. Bibollet-Ruche, J. E. Robinson, L. Morris, G. M. Shaw, D. C. Montefiori, and J. R. Mascola. 2008. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J. Virol. 82:11651-11668. - PMC - PubMed
-
- Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232-13252. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

