The XPA-binding domain of ERCC1 is required for nucleotide excision repair but not other DNA repair pathways
- PMID: 19940136
- PMCID: PMC2823511
- DOI: 10.1074/jbc.M109.067538
The XPA-binding domain of ERCC1 is required for nucleotide excision repair but not other DNA repair pathways
Abstract
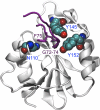
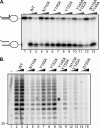
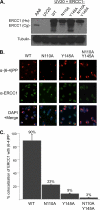
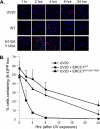
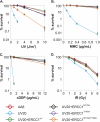
The endonuclease ERCC1-XPF incises the damaged strand of DNA 5' to a lesion during nucleotide excision repair (NER) and has additional, poorly characterized functions in interstrand cross-link repair, double-strand break repair, and homologous recombination. XPA, another key factor in NER, interacts with ERCC1 and recruits it to sites of damage. We identified ERCC1 residues that are critical for the interaction with XPA and assessed their importance for NER in vitro and in vivo. Mutation of two conserved residues (Asn-110 and Tyr-145) located in the XPA-binding site of ERCC1 dramatically affected NER but not nuclease activity on model DNA substrates. In ERCC1-deficient cells expressing ERCC1(N110A/Y145A), the nuclease was not recruited to sites of UV damage. The repair of UV-induced (6-4)photoproducts was severely impaired in these cells, and they were hypersensitive to UV irradiation. Remarkably, the ERCC1(N110A/Y145A) protein rescues the sensitivity of ERCC1-deficient cells to cross-linking agents. Our studies suggest that ERCC1-XPF engages in different repair pathways through specific protein-protein interactions and that these functions can be separated through the selective disruption of these interactions. We discuss the impact of these findings for understanding how ERCC1 contributes to resistance of tumor cells to therapeutic agents such as cisplatin.
Figures
Similar articles
-
Conformational determinants for the recruitment of ERCC1 by XPA in the nucleotide excision repair (NER) Pathway: structure and dynamics of the XPA binding motif.Biophys J. 2013 Jun 4;104(11):2503-11. doi: 10.1016/j.bpj.2013.04.023. Biophys J. 2013. PMID: 23746523 Free PMC article.
-
Functional and physical interactions between ERCC1 and MSH2 complexes for resistance to cis-diamminedichloroplatinum(II) in mammalian cells.DNA Repair (Amst). 2004 Feb 3;3(2):135-43. doi: 10.1016/j.dnarep.2003.10.005. DNA Repair (Amst). 2004. PMID: 14706347
-
Multiple DNA binding domains mediate the function of the ERCC1-XPF protein in nucleotide excision repair.J Biol Chem. 2012 Jun 22;287(26):21846-55. doi: 10.1074/jbc.M111.337899. Epub 2012 Apr 30. J Biol Chem. 2012. PMID: 22547097 Free PMC article.
-
DNA repair gets physical: mapping an XPA-binding site on ERCC1.DNA Repair (Amst). 2008 May 3;7(5):819-26. doi: 10.1016/j.dnarep.2008.01.018. Epub 2008 Mar 14. DNA Repair (Amst). 2008. PMID: 18343204 Free PMC article. Review.
-
Function and Interactions of ERCC1-XPF in DNA Damage Response.Molecules. 2018 Dec 5;23(12):3205. doi: 10.3390/molecules23123205. Molecules. 2018. PMID: 30563071 Free PMC article. Review.
Cited by
-
Mechanism and regulation of incisions during DNA interstrand cross-link repair.DNA Repair (Amst). 2014 Jul;19:135-42. doi: 10.1016/j.dnarep.2014.03.018. Epub 2014 Apr 24. DNA Repair (Amst). 2014. PMID: 24768452 Free PMC article. Review.
-
Suppression of isoprenylcysteine carboxylmethyltransferase compromises DNA damage repair.Life Sci Alliance. 2021 Oct 5;4(12):e202101144. doi: 10.26508/lsa.202101144. Print 2021 Dec. Life Sci Alliance. 2021. PMID: 34610973 Free PMC article.
-
The ERCC1 and ERCC4 (XPF) genes and gene products.Gene. 2015 Sep 15;569(2):153-61. doi: 10.1016/j.gene.2015.06.026. Epub 2015 Jun 12. Gene. 2015. PMID: 26074087 Free PMC article. Review.
-
Conformational determinants for the recruitment of ERCC1 by XPA in the nucleotide excision repair (NER) Pathway: structure and dynamics of the XPA binding motif.Biophys J. 2013 Jun 4;104(11):2503-11. doi: 10.1016/j.bpj.2013.04.023. Biophys J. 2013. PMID: 23746523 Free PMC article.
-
Unveiling Novel ERCC1-XPF Complex Inhibitors: Bridging the Gap from In Silico Exploration to Experimental Design.Int J Mol Sci. 2024 Jan 19;25(2):1246. doi: 10.3390/ijms25021246. Int J Mol Sci. 2024. PMID: 38279246 Free PMC article.
References
-
- Friedberg E. C., Walker G. C., Siede W., Wood R. D., Schultz R. A., Ellenberger T. (2005) DNA Repair and Mutagenesis, 2nd Ed., ASM Press, Washington, D. C.
-
- Gillet L. C., Schärer O. D. (2006) Chem. Rev. 106, 253–276 - PubMed
-
- Hanawalt P. C., Spivak G. (2008) Nat. Rev. Mol. Cell Biol. 9, 958–970 - PubMed
-
- Sugasawa K., Ng J. M., Masutani C., Iwai S., van der Spek P. J., Eker A. P., Hanaoka F., Bootsma D., Hoeijmakers J. H. (1998) Mol. Cell 2, 223–232 - PubMed
-
- Sugasawa K., Okuda Y., Saijo M., Nishi R., Matsuda N., Chu G., Mori T., Iwai S., Tanaka K., Tanaka K., Hanaoka F. (2005) Cell 121, 387–400 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

