Location of 3-hydroxyproline residues in collagen types I, II, III, and V/XI implies a role in fibril supramolecular assembly
- PMID: 19940144
- PMCID: PMC2807315
- DOI: 10.1074/jbc.M109.068726
Location of 3-hydroxyproline residues in collagen types I, II, III, and V/XI implies a role in fibril supramolecular assembly
Abstract
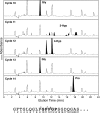
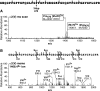
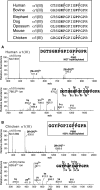
Collagen triple helices are stabilized by 4-hydroxyproline residues. No function is known for the much less common 3-hydroxyproline (3Hyp), although genetic defects inhibiting its formation cause recessive osteogenesis imperfecta. To help understand the pathogenesis, we used mass spectrometry to identify the sites and local sequence motifs of 3Hyp residues in fibril-forming collagens from normal human and bovine tissues. The results confirm a single, essentially fully occupied 3Hyp site (A1) at Pro(986) in A-clade chains alpha1(I), alpha1(II), and alpha2(V). Two partially modified sites (A2 and A3) were found at Pro(944) in alpha1(II) and alpha2(V) and Pro(707) in alpha2(I) and alpha2(V), which differed from A1 in sequence motif. Significantly, the distance between sites 2 and 3, 237 residues, is close to the collagen D-period (234 residues). A search for additional D-periodic 3Hyp sites revealed a fourth site (A4) at Pro(470) in alpha2(V), 237 residues N-terminal to site 3. In contrast, human and bovine type III collagen contained no 3Hyp at any site, despite a candidate proline residue and recognizable A1 sequence motif. A conserved histidine in mammalian alpha1(III) at A1 may have prevented 3-hydroxylation because this site in chicken type III was fully hydroxylated, and tyrosine replaced histidine. All three B-clade type V/XI collagen chains revealed the same three sites of 3Hyp but at different loci and sequence contexts from those in A-clade collagen chains. Two of these B-clade sites were spaced apart by 231 residues. From these and other observations we propose a fundamental role for 3Hyp residues in the ordered self-assembly of collagen supramolecular structures.
Figures






Similar articles
-
Insights on the evolution of prolyl 3-hydroxylation sites from comparative analysis of chicken and Xenopus fibrillar collagens.PLoS One. 2011 May 3;6(5):e19336. doi: 10.1371/journal.pone.0019336. PLoS One. 2011. PMID: 21559283 Free PMC article.
-
Evolutionary origins of C-terminal (GPP)n 3-hydroxyproline formation in vertebrate tendon collagen.PLoS One. 2014 Apr 2;9(4):e93467. doi: 10.1371/journal.pone.0093467. eCollection 2014. PLoS One. 2014. PMID: 24695516 Free PMC article.
-
A role for prolyl 3-hydroxylase 2 in post-translational modification of fibril-forming collagens.J Biol Chem. 2011 Sep 2;286(35):30662-30669. doi: 10.1074/jbc.M111.267906. Epub 2011 Jul 11. J Biol Chem. 2011. PMID: 21757687 Free PMC article.
-
Collagen prolyl 3-hydroxylation: a major role for a minor post-translational modification?Connect Tissue Res. 2013;54(4-5):245-51. doi: 10.3109/03008207.2013.800867. Epub 2013 Jun 21. Connect Tissue Res. 2013. PMID: 23772978 Free PMC article. Review.
-
The importance of proline residues in the structure, stability and susceptibility to proteolytic degradation of collagens.Amino Acids. 2008 Nov;35(4):703-10. doi: 10.1007/s00726-008-0073-2. Epub 2008 Apr 23. Amino Acids. 2008. PMID: 18431533 Review.
Cited by
-
Role of Glycosyltransferase 25 Domain 1 in Type I Collagen Glycosylation and Molecular Phenotypes.Biochemistry. 2019 Dec 17;58(50):5040-5051. doi: 10.1021/acs.biochem.8b00984. Epub 2019 Dec 5. Biochemistry. 2019. PMID: 31726007 Free PMC article.
-
Mechano-regulation of collagen biosynthesis in periodontal ligament.J Prosthodont Res. 2014 Oct;58(4):193-207. doi: 10.1016/j.jpor.2014.08.003. Epub 2014 Oct 11. J Prosthodont Res. 2014. PMID: 25311991 Free PMC article. Review.
-
Down-Regulation of Collagen Hydroxylation in Colorectal Liver Metastasis.Front Oncol. 2020 Sep 30;10:557737. doi: 10.3389/fonc.2020.557737. eCollection 2020. Front Oncol. 2020. PMID: 33117689 Free PMC article.
-
The multifaceted helical net of amphipathic alpha-helices; the next dimension of the helical peptide wheel.Sci Prog. 2024 Oct-Dec;107(4):368504241266357. doi: 10.1177/00368504241266357. Sci Prog. 2024. PMID: 39655381 Free PMC article.
-
Chaperoning osteogenesis: new protein-folding disease paradigms.Trends Cell Biol. 2011 Mar;21(3):168-76. doi: 10.1016/j.tcb.2010.11.007. Epub 2010 Dec 21. Trends Cell Biol. 2011. PMID: 21183349 Free PMC article. Review.
References
-
- Berg R. A., Prockop D. J. (1973) Biochem. Biophys. Res. Commun. 52, 115–120 - PubMed
-
- Ogle J. D., Arlinghaus R. B., Logan M. A. (1962) J. Biol. Chem. 237, 3667–3673 - PubMed
-
- Gryder R. M., Lamon M., Adams E. (1975) J. Biol. Chem. 250, 2470–2474 - PubMed
-
- Vranka J. A., Sakai L. Y., Bächinger H. P. (2004) J. Biol. Chem. 279, 23615–23621 - PubMed
-
- Tiainen P., Pasanen A., Sormunen R., Myllyharju J. (2008) J. Biol. Chem. 283, 19432–19439 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

