Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis
- PMID: 19940845
- PMCID: PMC2824244
- DOI: 10.1038/mi.2009.132
Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis
Abstract
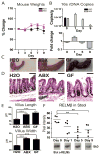
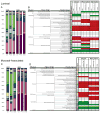
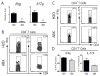
Despite widespread use of antibiotics, few studies have measured their effects on the burden or diversity of bacteria in the mammalian intestine. We developed an oral antibiotic treatment protocol and characterized its effects on murine intestinal bacterial communities and immune cell homeostasis. Antibiotic administration resulted in a 10-fold reduction in the amount of intestinal bacteria present and sequencing of 16S rDNA segments revealed significant temporal and spatial effects on luminal and mucosal-associated communities including reductions in luminal Firmicutes and mucosal-associated Lactobacillus species, and persistence of bacteria belonging to the Bacteroidetes and Proteobacteria phyla. Concurrently, antibiotic administration resulted in reduced RELM beta production, and reduced production of interferon-gamma and interleukin-17A by mucosal CD4(+) T lymphocytes. This comprehensive temporal and spatial metagenomic analyses will provide a resource and framework to test the influence of bacterial communities in murine models of human disease.
Figures
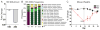

References
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI074878/AI/NIAID NIH HHS/United States
- DK50306/DK/NIDDK NIH HHS/United States
- P30 DK050306/DK/NIDDK NIH HHS/United States
- S10 RR024525/RR/NCRR NIH HHS/United States
- AI074878/AI/NIAID NIH HHS/United States
- T32 AI060516/AI/NIAID NIH HHS/United States
- T32 AI05528/AI/NIAID NIH HHS/United States
- R21 AI083480/AI/NIAID NIH HHS/United States
- R01 AI061570/AI/NIAID NIH HHS/United States
- T32 AI055428/AI/NIAID NIH HHS/United States
- R01 AI095466/AI/NIAID NIH HHS/United States
- S10RR024525/RR/NCRR NIH HHS/United States
- AI61570/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

