Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis
- PMID: 19941819
- PMCID: PMC2806949
- DOI: 10.1016/j.molcel.2009.09.039
Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis
Abstract
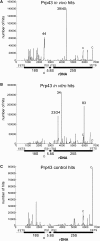
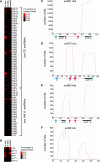
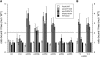
Yeast ribosome synthesis requires 19 different RNA helicases, but none of their pre-rRNA-binding sites were previously known, making their precise functions difficult to determine. Here we identify multiple binding sites for the helicase Prp43 in the 18S and 25S rRNA regions of pre-rRNAs, using UV crosslinking. Binding in 18S was predominantly within helix 44, close to the site of 18S 3' cleavage, in which Prp43 is functionally implicated. Four major binding sites were identified in 25S, including helix 34. In strains depleted of Prp43 or expressing only catalytic point mutants, six snoRNAs that guide modifications close to helix 34 accumulated on preribosomes, implicating Prp43 in their release, whereas other snoRNAs showed reduced preribosome association. Prp43 was crosslinked to snoRNAs that target sequences close to its binding sites, indicating direct interactions. We propose that Prp43 acts on preribosomal regions surrounding each binding site, with distinct functions at different locations.
Figures



Comment in
-
Helicase multitasking in ribosome assembly.Mol Cell. 2009 Nov 25;36(4):537-8. doi: 10.1016/j.molcel.2009.11.004. Mol Cell. 2009. PMID: 19941813
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

