Rep provides a second motor at the replisome to promote duplication of protein-bound DNA
- PMID: 19941825
- PMCID: PMC2807033
- DOI: 10.1016/j.molcel.2009.11.009
Rep provides a second motor at the replisome to promote duplication of protein-bound DNA
Abstract
Nucleoprotein complexes present challenges to genome stability by acting as potent blocks to replication. One attractive model of how such conflicts are resolved is direct targeting of blocked forks by helicases with the ability to displace the blocking protein-DNA complex. We show that Rep and UvrD each promote movement of E. coli replisomes blocked by nucleoprotein complexes in vitro, that such an activity is required to clear protein blocks (primarily transcription complexes) in vivo, and that a polarity of translocation opposite that of the replicative helicase is critical for this activity. However, these two helicases are not equivalent. Rep but not UvrD interacts physically and functionally with the replicative helicase. In contrast, UvrD likely provides a general means of protein-DNA complex turnover during replication, repair, and recombination. Rep and UvrD therefore provide two contrasting solutions as to how organisms may promote replication of protein-bound DNA.
Figures
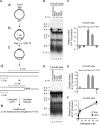
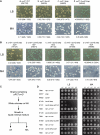


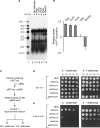
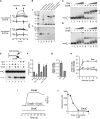
References
-
- Aguilera A., Gómez-González B. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008;9:204–217. - PubMed
-
- Bedinger P., Hochstrasser M., Jongeneel C.V., Alberts B.M. Properties of the T4 bacteriophage DNA replication apparatus: the T4 dda DNA helicase is required to pass a bound RNA polymerase molecule. Cell. 1983;34:115–123. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/C008316/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/G005915/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 077368/Z/05/Z/WT_/Wellcome Trust/United Kingdom
- G0800970/MRC_/Medical Research Council/United Kingdom
- BB/E0020690/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

