Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy
- PMID: 19941826
- PMCID: PMC2784893
- DOI: 10.1016/j.molcel.2009.11.001
Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy
Abstract
GATA factors interact with simple DNA motifs (WGATAR) to regulate critical processes, including hematopoiesis, but very few WGATAR motifs are occupied in genomes. Given the rudimentary knowledge of mechanisms underlying this restriction and how GATA factors establish genetic networks, we used ChIP-seq to define GATA-1 and GATA-2 occupancy genome-wide in erythroid cells. Coupled with genetic complementation analysis and transcriptional profiling, these studies revealed a rich collection of targets containing a characteristic binding motif of greater complexity than WGATAR. GATA factors occupied loci encoding multiple components of the Scl/TAL1 complex, a master regulator of hematopoiesis and leukemogenic target. Mechanistic analyses provided evidence for crossregulatory and autoregulatory interactions among components of this complex, including GATA-2 induction of the hematopoietic corepressor ETO-2 and an ETO-2-negative autoregulatory loop. These results establish fundamental principles underlying GATA factor mechanisms in chromatin and illustrate a complex network of considerable importance for the control of hematopoiesis.
Figures


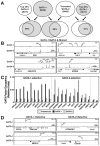



References
-
- Aplan PD, Lombardi DP, Reaman GH, Sather HN, Hammond GD, Kirsch IR. Involvement of the putative hematopoietic transcription factor SCL in T- cell acute lymphoblastic leukemia. Blood. 1992;79:1327–1333. - PubMed
-
- Bailey TL, Elkan C. Unsupervised learning of multiple motifs in biopolymers using EM. Machine Learning. 1995;21:51–80.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- U54 HG004558/HG/NHGRI NIH HHS/United States
- R01 DK050107/DK/NIDDK NIH HHS/United States
- 1U54HG004558/HG/NHGRI NIH HHS/United States
- DK50107/DK/NIDDK NIH HHS/United States
- R56 DK068634/DK/NIDDK NIH HHS/United States
- R29 HL055337/HL/NHLBI NIH HHS/United States
- DK068634/DK/NIDDK NIH HHS/United States
- HL55337/HL/NHLBI NIH HHS/United States
- R01 HG003747/HG/NHGRI NIH HHS/United States
- R01 HL055337/HL/NHLBI NIH HHS/United States
- R01 DK068634/DK/NIDDK NIH HHS/United States
- HG003747/HG/NHGRI NIH HHS/United States
- R37 DK050107/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

