PCNA is required for initiation of recombination-associated DNA synthesis by DNA polymerase delta
- PMID: 19941829
- PMCID: PMC2784891
- DOI: 10.1016/j.molcel.2009.09.036
PCNA is required for initiation of recombination-associated DNA synthesis by DNA polymerase delta
Abstract
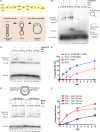
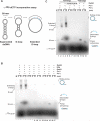
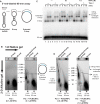
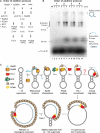
Genetic recombination ensures proper chromosome segregation during meiosis and is essential for genome stability and tumor suppression. DNA synthesis after Rad51-mediated DNA strand invasion is a crucial step during recombination. PCNA is known as the processivity clamp for DNA polymerases. Here, we report the surprising observation that PCNA is specifically required to initiate recombination-associated DNA synthesis in the extension of the 3' end of the invading strand in a D loop. We show using a reconstituted system of yeast Rad51, Rad54, RPA, PCNA, RFC, and DNA polymerase delta that loading of PCNA by RFC targets DNA polymerase delta to the D loop formed by Rad51 protein, allowing efficient utilization of the invading 3' end and processive DNA synthesis. We conclude that PCNA has a specific role in the initiation of recombination-associated DNA synthesis and that DNA polymerase delta promotes recombination-associated DNA synthesis.
Figures
Similar articles
-
RAD54 controls access to the invading 3'-OH end after RAD51-mediated DNA strand invasion in homologous recombination in Saccharomyces cerevisiae.Nucleic Acids Res. 2009 Feb;37(2):638-46. doi: 10.1093/nar/gkn980. Epub 2008 Dec 11. Nucleic Acids Res. 2009. PMID: 19074197 Free PMC article.
-
Reconstitution of recombination-associated DNA synthesis with human proteins.Nucleic Acids Res. 2013 May;41(9):4913-25. doi: 10.1093/nar/gkt192. Epub 2013 Mar 27. Nucleic Acids Res. 2013. PMID: 23535143 Free PMC article.
-
The Pif1 helicase is actively inhibited during meiotic recombination which restrains gene conversion tract length.Nucleic Acids Res. 2021 May 7;49(8):4522-4533. doi: 10.1093/nar/gkab232. Nucleic Acids Res. 2021. PMID: 33823531 Free PMC article.
-
Functions of the Snf2/Swi2 family Rad54 motor protein in homologous recombination.Biochim Biophys Acta. 2011 Sep;1809(9):509-23. doi: 10.1016/j.bbagrm.2011.06.006. Epub 2011 Jun 16. Biochim Biophys Acta. 2011. PMID: 21704205 Free PMC article. Review.
-
DNA polymerase epsilon: the latest member in the family of mammalian DNA polymerases.Bioessays. 1990 Nov;12(11):533-6. doi: 10.1002/bies.950121106. Bioessays. 1990. PMID: 2085320 Review.
Cited by
-
Mismatch repair during homologous and homeologous recombination.Cold Spring Harb Perspect Biol. 2015 Mar 2;7(3):a022657. doi: 10.1101/cshperspect.a022657. Cold Spring Harb Perspect Biol. 2015. PMID: 25731766 Free PMC article. Review.
-
Rad51 and Rad54 promote noncrossover recombination between centromere repeats on the same chromatid to prevent isochromosome formation.Nucleic Acids Res. 2016 Dec 15;44(22):10744-10757. doi: 10.1093/nar/gkw874. Epub 2016 Oct 3. Nucleic Acids Res. 2016. PMID: 27697832 Free PMC article.
-
A novel function of CRL4(Cdt2): regulation of the subunit structure of DNA polymerase δ in response to DNA damage and during the S phase.J Biol Chem. 2013 Oct 11;288(41):29550-61. doi: 10.1074/jbc.M113.490466. Epub 2013 Aug 2. J Biol Chem. 2013. PMID: 23913683 Free PMC article.
-
Recombinational Repair of Nuclease-Generated Mitotic Double-Strand Breaks with Different End Structures in Yeast.G3 (Bethesda). 2020 Oct 5;10(10):3821-3829. doi: 10.1534/g3.120.401603. G3 (Bethesda). 2020. PMID: 32826304 Free PMC article.
-
Revisiting the Function of p21CDKN1A in DNA Repair: The Influence of Protein Interactions and Stability.Int J Mol Sci. 2022 Jun 24;23(13):7058. doi: 10.3390/ijms23137058. Int J Mol Sci. 2022. PMID: 35806061 Free PMC article. Review.
References
-
- Amitani I, Baskin RJ, Kowalczykowski SC. Visualization of Rad54, a chromatin remodeling protein, translocating on single DNA molecules. Mol Cell. 2006;23:143–148. - PubMed
-
- Bugreev DV, Hanaoka F, Mazin AV. Rad54 dissociates homologous recombination intermediates by branch migration. Nature Struct Mol Biol. 2007;14:746–753. - PubMed
-
- Dupaigne P, Le Breton C, Fabre F, Giangloff S, Le Cam E, Veaute X. The Srs2 helicase activity is stimulated by Rad51 filaments on dsDNA: Implications for crossover incidence during mitotic recombination. Mol. Cell. 2008;29:243–254. - PubMed
-
- Eggler AL, Inman RB, Cox MM. The Rad51-dependent pairing of long DNA substrates is stabilized by replication protein. A. J. Biol. Chem. 2002;277:39280–39288. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

