The balance between pre- and post-transfer editing in tRNA synthetases
- PMID: 19941860
- PMCID: PMC2859721
- DOI: 10.1016/j.febslet.2009.11.071
The balance between pre- and post-transfer editing in tRNA synthetases
Abstract
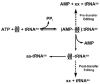
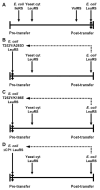
The fidelity of tRNA aminoacylation is dependent in part on amino acid editing mechanisms. A hydrolytic activity that clears mischarged tRNAs typically resides in an active site on the tRNA synthetase that is distinct from its synthetic aminoacylation active site. A second pre-transfer editing pathway that hydrolyzes the tRNA synthetase aminoacyl adenylate intermediate can also be activated. Pre- and post-transfer editing activities can co-exist within a single tRNA synthetase resulting in a redundancy of fidelity mechanisms. However, in most cases one pathway appears to dominate, but when compromised, the secondary pathway can be activated to suppress tRNA synthetase infidelities.
Figures

Similar articles
-
Pre-transfer editing by class II prolyl-tRNA synthetase: role of aminoacylation active site in "selective release" of noncognate amino acids.J Biol Chem. 2006 Sep 22;281(38):27862-72. doi: 10.1074/jbc.M605856200. Epub 2006 Jul 24. J Biol Chem. 2006. PMID: 16864571
-
Transfer RNA modulates the editing mechanism used by class II prolyl-tRNA synthetase.J Biol Chem. 2008 Mar 14;283(11):7128-34. doi: 10.1074/jbc.M709902200. Epub 2008 Jan 7. J Biol Chem. 2008. PMID: 18180290
-
Determinants for tRNA-dependent pretransfer editing in the synthetic site of isoleucyl-tRNA synthetase.Biochemistry. 2014 Oct 7;53(39):6189-98. doi: 10.1021/bi5007699. Epub 2014 Sep 23. Biochemistry. 2014. PMID: 25207837 Free PMC article.
-
Trans-editing by aminoacyl-tRNA synthetase-like editing domains.Enzymes. 2020;48:69-115. doi: 10.1016/bs.enz.2020.07.002. Epub 2020 Sep 8. Enzymes. 2020. PMID: 33837712 Review.
-
Synthetic and editing reactions of aminoacyl-tRNA synthetases using cognate and non-cognate amino acid substrates.Methods. 2017 Jan 15;113:13-26. doi: 10.1016/j.ymeth.2016.09.015. Epub 2016 Oct 3. Methods. 2017. PMID: 27713080 Review.
Cited by
-
Genome-wide screening reveals metabolic regulation of stop-codon readthrough by cyclic AMP.Nucleic Acids Res. 2023 Oct 13;51(18):9905-9919. doi: 10.1093/nar/gkad725. Nucleic Acids Res. 2023. PMID: 37670559 Free PMC article.
-
Backbone Brackets and Arginine Tweezers delineate Class I and Class II aminoacyl tRNA synthetases.PLoS Comput Biol. 2018 Apr 16;14(4):e1006101. doi: 10.1371/journal.pcbi.1006101. eCollection 2018 Apr. PLoS Comput Biol. 2018. PMID: 29659563 Free PMC article.
-
Do anticodons of misacylated tRNAs preferentially mismatch codons coding for the misloaded amino acid?BMC Mol Biol. 2010 May 28;11:41. doi: 10.1186/1471-2199-11-41. BMC Mol Biol. 2010. PMID: 20509917 Free PMC article.
-
Cross-editing by a tRNA synthetase allows vertebrates to abundantly express mischargeable tRNA without causing mistranslation.Nucleic Acids Res. 2020 Jul 9;48(12):6445-6457. doi: 10.1093/nar/gkaa469. Nucleic Acids Res. 2020. PMID: 32484512 Free PMC article.
-
p53-Dependent DNA damage response sensitive to editing-defective tRNA synthetase in zebrafish.Proc Natl Acad Sci U S A. 2016 Jul 26;113(30):8460-5. doi: 10.1073/pnas.1608139113. Epub 2016 Jul 8. Proc Natl Acad Sci U S A. 2016. PMID: 27402763 Free PMC article.
References
-
- Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, Davisson MT, Sundberg JP, Schimmel P, Ackerman SL. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. - PubMed
-
- Hoagland MB, Keller EB, Zamecnik PC. Enzymatic carboxyl activation of amino acids. J Biol Chem. 1956;218:345–358. - PubMed
-
- Pauling L, editor. Festschrift Arthur Stöll Siebzigsten Geburtstag. Birkhauser Verlag; Basel: 1958. p. 597.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

