Origin and diversification of basic-helix-loop-helix proteins in plants
- PMID: 19942615
- PMCID: PMC2839125
- DOI: 10.1093/molbev/msp288
Origin and diversification of basic-helix-loop-helix proteins in plants
Abstract
Basic helix-loop-helix (bHLH) proteins are a class of transcription factors found throughout eukaryotic organisms. Classification of the complete sets of bHLH proteins in the sequenced genomes of Arabidopsis thaliana and Oryza sativa (rice) has defined the diversity of these proteins among flowering plants. However, the evolutionary relationships of different plant bHLH groups and the diversity of bHLH proteins in more ancestral groups of plants are currently unknown. In this study, we use whole-genome sequences from nine species of land plants and algae to define the relationships between these proteins in plants. We show that few (less than 5) bHLH proteins are encoded in the genomes of chlorophytes and red algae. In contrast, many bHLH proteins (100-170) are encoded in the genomes of land plants (embryophytes). Phylogenetic analyses suggest that plant bHLH proteins are monophyletic and constitute 26 subfamilies. Twenty of these subfamilies existed in the common ancestors of extant mosses and vascular plants, whereas six further subfamilies evolved among the vascular plants. In addition to the conserved bHLH domains, most subfamilies are characterized by the presence of highly conserved short amino acid motifs. We conclude that much of the diversity of plant bHLH proteins was established in early land plants, over 440 million years ago.
Figures

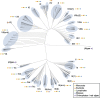


Comment in
-
Early evolution of bHLH proteins in plants.Plant Signal Behav. 2010 Jul;5(7):911-2. doi: 10.4161/psb.5.7.12100. Epub 2010 Jul 1. Plant Signal Behav. 2010. PMID: 20523129 Free PMC article.
References
-
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. - PubMed
-
- Anantharaman V, Koonin EV, Aravind L. Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J Mol Biol. 2001;307:1271–1292. - PubMed
-
- Armbrust EV, Berges JA, Bowler C, et al. (45 co-authors) The genome of the diatom Thalassiosira Pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

