Production of clastogenic DNA precursors by the nucleotide metabolism in Escherichia coli
- PMID: 19943897
- PMCID: PMC4433007
- DOI: 10.1111/j.1365-2958.2009.06994.x
Production of clastogenic DNA precursors by the nucleotide metabolism in Escherichia coli
Abstract
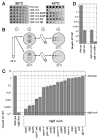
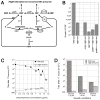
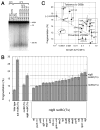
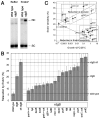
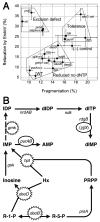
RdgB is a bacterial dNTPase with a strong in vitro preference for non-canonical DNA precursors dHapTP, dXTP and dITP that contain deaminated or aminogroup-modified purines. Utilization of these nucleotides by replisomes in rdgB mutants of Escherichia coli produces modified DNA, on which EndoV nicking near the base analogues initiates excision repair. Some EndoV-initiated excision events cause chromosomal fragmentation, which becomes inhibitory if recombinational repair is also inactivated (the rdgB recA co-inhibition). To reveal the sources and the identities of the non-canonical DNA precursors, intercepted by RdgB in E. coli, we characterized 17 suppressors of the rdgB recA co-inhibition. Ten suppressors affect genes of the RNA/DNA precursor metabolism, identifying the source of non-canonical DNA precursors. Comparing chromosomal fragmentation with the density of EndoV-recognized DNA modifications distinguishes three mechanisms of suppression: (i) reduction of the non-canonical dNTP production, (ii) inhibition of the base analogue excision from DNA and (iii) enhancement of the cell tolerance to chromosomal fragmentation. The suppressor analysis suggests IMP as the key intermediate in the synthesis of the clastogenic DNA precursor, most likely dITP.
Figures

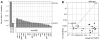
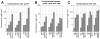
Similar articles
-
RdgB acts to avoid chromosome fragmentation in Escherichia coli.Mol Microbiol. 2003 Jun;48(6):1711-25. doi: 10.1046/j.1365-2958.2003.03540.x. Mol Microbiol. 2003. PMID: 12791149
-
The mutT defect does not elevate chromosomal fragmentation in Escherichia coli because of the surprisingly low levels of MutM/MutY-recognized DNA modifications.J Bacteriol. 2007 Oct;189(19):6976-88. doi: 10.1128/JB.00776-07. Epub 2007 Jul 6. J Bacteriol. 2007. PMID: 17616589 Free PMC article.
-
Hypoxanthine incorporation is nonmutagenic in Escherichia coli.J Bacteriol. 2006 Sep;188(18):6553-60. doi: 10.1128/JB.00447-06. J Bacteriol. 2006. PMID: 16952947 Free PMC article.
-
Recombinational DNA repair: the ignored repair systems.Bioessays. 2004 Dec;26(12):1322-6. doi: 10.1002/bies.20109. Bioessays. 2004. PMID: 15551273 Review.
-
RecA: Regulation and Mechanism of a Molecular Search Engine.Trends Biochem Sci. 2016 Jun;41(6):491-507. doi: 10.1016/j.tibs.2016.04.002. Epub 2016 May 4. Trends Biochem Sci. 2016. PMID: 27156117 Free PMC article. Review.
Cited by
-
Genomewide phenotypic analysis of growth, cell morphogenesis, and cell cycle events in Escherichia coli.Mol Syst Biol. 2018 Jun 25;14(6):e7573. doi: 10.15252/msb.20177573. Mol Syst Biol. 2018. PMID: 29941428 Free PMC article.
-
Measuring deaminated nucleotide surveillance enzyme ITPA activity with an ATP-releasing nucleotide chimera.Nucleic Acids Res. 2017 Nov 16;45(20):11515-11524. doi: 10.1093/nar/gkx774. Nucleic Acids Res. 2017. PMID: 29036687 Free PMC article.
-
Pivotal role of inosine triphosphate pyrophosphatase in maintaining genome stability and the prevention of apoptosis in human cells.PLoS One. 2012;7(2):e32313. doi: 10.1371/journal.pone.0032313. Epub 2012 Feb 27. PLoS One. 2012. PMID: 22384212 Free PMC article.
-
Cyanide enhances hydrogen peroxide toxicity by recruiting endogenous iron to trigger catastrophic chromosomal fragmentation.Mol Microbiol. 2015 Apr;96(2):349-67. doi: 10.1111/mmi.12938. Epub 2015 Feb 18. Mol Microbiol. 2015. PMID: 25598241 Free PMC article.
-
Replication Fork Breakage and Restart in Escherichia coli.Microbiol Mol Biol Rev. 2018 Jun 13;82(3):e00013-18. doi: 10.1128/MMBR.00013-18. Print 2018 Sep. Microbiol Mol Biol Rev. 2018. PMID: 29898897 Free PMC article. Review.
References
-
- Abdul-Masih MT, Bessman MJ. Biochemical studies on the mutagen, 6-N-hydroxylaminopurine. Synthesis of the deoxynucleoside triphosphate and its incorporation into DNA in vitro. J Biol Chem. 1986;261:2020–2026. - PubMed
-
- Birnboim HC. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 1983;100:243–255. - PubMed
-
- Bradshaw JS, Kuzminov A. RdgB acts to avoid chromosome fragmentation in Escherichia coli. Mol Microbiol. 2003;48:1711–1725. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

