Spectral affinity in protein networks
- PMID: 19943959
- PMCID: PMC2797010
- DOI: 10.1186/1752-0509-3-112
Spectral affinity in protein networks
Abstract
Background: Protein-protein interaction (PPI) networks enable us to better understand the functional organization of the proteome. We can learn a lot about a particular protein by querying its neighborhood in a PPI network to find proteins with similar function. A spectral approach that considers random walks between nodes of interest is particularly useful in evaluating closeness in PPI networks. Spectral measures of closeness are more robust to noise in the data and are more precise than simpler methods based on edge density and shortest path length.
Results: We develop a novel affinity measure for pairs of proteins in PPI networks, which uses personalized PageRank, a random walk based method used in context-sensitive search on the Web. Our measure of closeness, which we call PageRank Affinity, is proportional to the number of times the smaller-degree protein is visited in a random walk that restarts at the larger-degree protein. PageRank considers paths of all lengths in a network, therefore PageRank Affinity is a precise measure that is robust to noise in the data. PageRank Affinity is also provably related to cluster co-membership, making it a meaningful measure. In our experiments on protein networks we find that our measure is better at predicting co-complex membership and finding functionally related proteins than other commonly used measures of closeness. Moreover, our experiments indicate that PageRank Affinity is very resilient to noise in the network. In addition, based on our method we build a tool that quickly finds nodes closest to a queried protein in any protein network, and easily scales to much larger biological networks.
Conclusion: We define a meaningful way to assess the closeness of two proteins in a PPI network, and show that our closeness measure is more biologically significant than other commonly used methods. We also develop a tool, accessible at http://xialab.bu.edu/resources/pnns, that allows the user to quickly find nodes closest to a queried vertex in any protein network available from BioGRID or specified by the user.
Figures

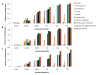
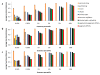

Similar articles
-
Finding local communities in protein networks.BMC Bioinformatics. 2009 Sep 18;10:297. doi: 10.1186/1471-2105-10-297. BMC Bioinformatics. 2009. PMID: 19765306 Free PMC article.
-
Identification of Protein Complexes Using Weighted PageRank-Nibble Algorithm and Core-Attachment Structure.IEEE/ACM Trans Comput Biol Bioinform. 2015 Jan-Feb;12(1):179-92. doi: 10.1109/TCBB.2014.2343954. IEEE/ACM Trans Comput Biol Bioinform. 2015. PMID: 26357088
-
Constructing multilayer PPI networks based on homologous proteins and integrating multiple PageRank to identify essential proteins.BMC Bioinformatics. 2025 Mar 10;26(1):80. doi: 10.1186/s12859-025-06093-5. BMC Bioinformatics. 2025. PMID: 40059137 Free PMC article.
-
Discerning molecular interactions: A comprehensive review on biomolecular interaction databases and network analysis tools.Gene. 2018 Feb 5;642:84-94. doi: 10.1016/j.gene.2017.11.028. Epub 2017 Nov 10. Gene. 2018. PMID: 29129810 Review.
-
Construction and contextualization approaches for protein-protein interaction networks.Comput Struct Biotechnol J. 2022 Jun 18;20:3280-3290. doi: 10.1016/j.csbj.2022.06.040. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35832626 Free PMC article. Review.
Cited by
-
It's the machine that matters: Predicting gene function and phenotype from protein networks.J Proteomics. 2010 Oct 10;73(11):2277-89. doi: 10.1016/j.jprot.2010.07.005. Epub 2010 Jul 15. J Proteomics. 2010. PMID: 20637909 Free PMC article. Review.
-
Information flow in interaction networks II: channels, path lengths, and potentials.J Comput Biol. 2012 Apr;19(4):379-403. doi: 10.1089/cmb.2010.0228. Epub 2012 Mar 12. J Comput Biol. 2012. PMID: 22409812 Free PMC article.
-
A single source k-shortest paths algorithm to infer regulatory pathways in a gene network.Bioinformatics. 2012 Jun 15;28(12):i49-58. doi: 10.1093/bioinformatics/bts212. Bioinformatics. 2012. PMID: 22689778 Free PMC article.
-
Inferring hidden causal relations between pathway members using reduced Google matrix of directed biological networks.PLoS One. 2018 Jan 25;13(1):e0190812. doi: 10.1371/journal.pone.0190812. eCollection 2018. PLoS One. 2018. PMID: 29370181 Free PMC article.
-
Network propagation: a universal amplifier of genetic associations.Nat Rev Genet. 2017 Sep;18(9):551-562. doi: 10.1038/nrg.2017.38. Epub 2017 Jun 12. Nat Rev Genet. 2017. PMID: 28607512 Review.
References
-
- Fowler J. Legislative cosponsorship networks in the US House and Senate. Social Networks. 2006;28:454–465. doi: 10.1016/j.socnet.2005.11.003. - DOI
-
- Gibson D, Kleinberg J, Raghavan P. Inferring Web communities from link topology. Proc ACM Conf on Hypertext and Hypermedia. 1998. pp. 225–234.
-
- Kumar R, Raghavan P, Rajagopalan S, Tomkins A. Trawling the Web for emerging cyber-communities. Computer Networks. 1999;31:1481–1493. doi: 10.1016/S1389-1286(99)00040-7. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

