Stereochemical mechanisms of tRNA methyltransferases
- PMID: 19944101
- PMCID: PMC2797553
- DOI: 10.1016/j.febslet.2009.11.075
Stereochemical mechanisms of tRNA methyltransferases
Abstract
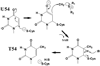
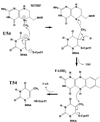
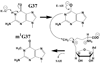
Methylation of tRNA on the four canonical bases adds structural complexity to the molecule, and improves decoding specificity and efficiency. While many tRNA methylases are known, detailed insight into the catalytic mechanism is only available in a few cases. Of interest among all tRNA methylases is the structural basis for nucleotide selection, by which the specificity is limited to a single site, or broadened to multiple sites. General themes in catalysis include the basis for rate acceleration at highly diverse nucleophilic centers for methyl transfer, using S-adenosylmethionine as a cofactor. Studies of tRNA methylases have also yielded insights into molecular evolution, particularly in the case of enzymes that recognize distinct structures to perform identical reactions at the same target nucleotide.
Figures
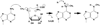

References
-
- Bjork GR, Ericson JU, Gustafsson CE, Hagervall TG, Jonsson YH, Wikstrom PM. Transfer RNA modification. Annu Rev Biochem. 1987;56:263–287. - PubMed
-
- Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell. 2006;21:87–96. - PubMed
-
- Cantoni GL. Biological methylation: selected aspects. Annu Rev Biochem. 1975;44:435–451. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

